Production of infectious hepatitis C virus in tissue culture from a cloned viral genome - PubMed (original) (raw)
doi: 10.1038/nm1268. Epub 2005 Jun 12.
Thomas Pietschmann, Takanobu Kato, Tomoko Date, Michiko Miyamoto, Zijiang Zhao, Krishna Murthy, Anja Habermann, Hans-Georg Kräusslich, Masashi Mizokami, Ralf Bartenschlager, T Jake Liang
Affiliations
- PMID: 15951748
- PMCID: PMC2918402
- DOI: 10.1038/nm1268
Production of infectious hepatitis C virus in tissue culture from a cloned viral genome
Takaji Wakita et al. Nat Med. 2005 Jul.
Erratum in
- Nat Med. 2005 Aug;11(8):905
Abstract
Hepatitis C virus (HCV) infection causes chronic liver diseases and is a global public health problem. Detailed analyses of HCV have been hampered by the lack of viral culture systems. Subgenomic replicons of the JFH1 genotype 2a strain cloned from an individual with fulminant hepatitis replicate efficiently in cell culture. Here we show that the JFH1 genome replicates efficiently and supports secretion of viral particles after transfection into a human hepatoma cell line (Huh7). Particles have a density of about 1.15-1.17 g/ml and a spherical morphology with an average diameter of about 55 nm. Secreted virus is infectious for Huh7 cells and infectivity can be neutralized by CD81-specific antibodies and by immunoglobulins from chronically infected individuals. The cell culture-generated HCV is infectious for chimpanzee. This system provides a powerful tool for studying the viral life cycle and developing antiviral strategies.
Conflict of interest statement
Competing Interests Statement: The authors declare that they have no competing financial interests.
Figures
Figure 1
Transient replication of JFH1 RNA in transfected Huh7 cells. (a) Organization of the full-length HCV construct pJFH1. Open reading frames (thick boxes) are flanked by the 5′- and 3′-UTRs (thin boxes). T7, T7 RNA polymerase promoter; GDD, active-site motif of NS5B polymerase; XbaI, restriction site. (b) Northern blot analysis of total RNA prepared from cells transfected with full-length JFH1 and JFH1/GND RNA. Control RNA, given numbers of synthetic HCV RNA; Huh7, RNA isolated from naive cells. Arrowheads indicate full-length HCV RNA and 28S ribosomal RNA (28S). Upper panel, northern blot; lower panel, ethidium bromide staining. (c) Western blot analysis of transfected cells for HCV proteins NS5A, NS3 and core. Lysates of SGR/JFH1-RNA or pEF/Core DNA– transfected Huh7 cells and naive Huh7 cells served as positive and negative controls, respectively. (d) Immunofluorescence assay of cells fixed 72 h after transfection with JFH1 or JFH1/ΔE1-E2 RNA. Magnification, ×200. (e) HCV core protein secretion into culture medium. HCV RNAs were transfected into Huh7 cells, culture media were harvested at given time points, and HCV core protein concentrations were determined.
Figure 2
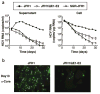
Time course of HCV RNA replication and core protein expression in transfected cells. (a) Levels of HCV RNA in transfected cells (right) and corresponding culture supernatants (left). Huh7 cells were transfected with given HCV RNAs and passaged up to 30 d. Viral RNA was determined 2, 5, 8, 12, 15, 18, 21, 24, 27 and 30 d after transfection. (b) Immunofluorescence assay of HCV core protein in passaged cells transfected with JFH1 and JFH1/ΔE1-E2 RNA. Transfected cells were harvested 1 d after the fourth passage (day 13 after transfection). Magnification, ×100.
Figure 3
Density gradient and electron microscope analysis of recombinant HCV particles. (a,b) Co-sedimentation of viral RNA and structural proteins. (a) Concentrated culture medium collected from JFH1/E2HA RNA–transfected cells was fractionated using a 10–60% sucrose density gradient. HCV RNA titer in each fraction was determined. (b) Density gradient fractions were further concentrated and analyzed by western blotting for core, E1 or E2-hemagglutinin. P, cell lysate prepared from JFH1/E2HA RNA–transfected Huh7 cells; N, cell lysate from untransfected Huh7 cells. Arrowheads indicate positions of HCV proteins. (c–f) Electron micrograph of spherical structures shown by immunogold labeling. Grids were incubated with a concentrated JFH1 virus stock and then with the E2 monoclonal antibody CBH5 (ref. 14). Bound antibodies were detected with Protein A coupled to gold particles 10 nm in diameter. (c–e) Three representative examples showing the same structure. (f) Control grid coated with concentrated cell-free supernatant derived from mock-transfected cells. In rare cases, we observed gold particles attached to unstructured protein aggregates. Scale bar, 50 nm.
Figure 4
Infectivity of viral particles and neutralization of infection. (a, insert) Immunofluorescence analysis of cells infected with viral particles. Huh7 cells inoculated with virus-containing medium were stained simultaneously for core (left) and NS5A (right). (a) Cell lines specified on the bottom were also inoculated with a 30-fold concentrated supernatant from full-length JFH1 RNA– or JFH1/ΔE1-E2 RNA– transfected cells (Sup). In some experiments, culture supernatant of nontransfected cells was used (EP−), or culture supernatant was irradiated with ultraviolet light before inoculation of cells (UV+). We stained cells 2 d after infection for core protein and counted positive cells. (b) Comparison of infectivity of culture supernatant from JFH1 RNA– and JFH1/ΔE1-E2 RNA– transfected cells. Supernatants containing about 108 RNA copies/ml were used for inoculation of Huh7 cells and amounts of cell-associated RNA were determined 0, 12, 24 and 48 h after inoculation. (c) Inhibition of infection by CD81-specific antibody. We used 20-fold concentrated supernatants from cells transfected with given genomes for infection of Huh7 cells in the presence of a CD81-specific (α-CD81, black bars) or a control antibody (Ctrl Ab, gray bars), or in the absence of antibody (Ab(−), white bars). Inoculated cells were analyzed by RTD-PCR. (d) Production of infectious HCV particles carrying the firefly luciferase reporter gene and neutralization of infection by sera from infected individuals. Upper panel, schematic representation of Luc-JFH1 construct with luciferase (Luc) reporter gene (Supplementary Fig. 6 online). E-I, EMCV-IRES. Bottom left panel, neutralization of Luc-JFH1 virus by sera from infected individuals. Culture supernatants containing Luc-JFH1 viral particles were mixed with given dilutions of sera from healthy donor (Control), or sera from individuals infected with given genotypes (lanes 1–9). Results of CD81-specific antibody neutralization are shown in the right; black bar, 2 μg/ml; gray bar, 0.4 μg/ml; white bar, 0.08 μg/ml. Luciferase activity was determined 72 h later and is expressed relative to the values obtained with control serum A. Bottom right panel, neutralization by immunoglobulins purified from infected individuals' sera. Luc-JFH1 virus–containing supernatant was mixed with 2 mg total protein contained in control serum B or patient serum 3 (serum, black bars). Addition of this amount of serum protein is equivalent to a final serum dilution of approximately 1:20. Alternatively, virus preparation was mixed with 2 mg protein of the same sera depleted of immunoglobulins (Serum Ig(−), open bars), or 130 μg of corresponding purified immunoglobulins (Ig, gray bars). Infectivity is expressed relative to control serum B.
Figure 5. In vivo
infectivity of JFH1 virus produced In tissue culture. Chimpanzee X0215 was first inoculated with 1 ml of the undiluted culture medium from mock-transfected Huh7 cells (Ctrl). We re-inoculated the chimpanzee 6 weeks later with 1 ml of a 104 dilution of culture medium from full-length JFH1 RNA-transfected cells (104). After 6 more weeks, we repeated inoculatlon with 1 ml of a 103 dilution (103). The course of infection is shown with arrows indicating the three inoculations. HCV RNA (copies/ml) and ALT (IU/L) levels are plotted; HCV-specific, HCV RNA and liver biopsy results are shown above the graph.
Similar articles
- Construction of the Vero cell culture system that can produce infectious HCV particles.
Guo J, Yan R, Xu G, Li W, Zheng C. Guo J, et al. Mol Biol Rep. 2009 Jan;36(1):111-20. doi: 10.1007/s11033-007-9158-3. Epub 2007 Oct 25. Mol Biol Rep. 2009. PMID: 17960493 - Cell culture and infection system for hepatitis C virus.
Kato T, Date T, Murayama A, Morikawa K, Akazawa D, Wakita T. Kato T, et al. Nat Protoc. 2006;1(5):2334-9. doi: 10.1038/nprot.2006.395. Nat Protoc. 2006. PMID: 17406476 - Cutting the gordian knot-development and biological relevance of hepatitis C virus cell culture systems.
Gottwein JM, Bukh J. Gottwein JM, et al. Adv Virus Res. 2008;71:51-133. doi: 10.1016/S0065-3527(08)00002-X. Adv Virus Res. 2008. PMID: 18585527 Review. - Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells.
Cai Z, Zhang C, Chang KS, Jiang J, Ahn BC, Wakita T, Liang TJ, Luo G. Cai Z, et al. J Virol. 2005 Nov;79(22):13963-73. doi: 10.1128/JVI.79.22.13963-13973.2005. J Virol. 2005. PMID: 16254332 Free PMC article. - Novel cell culture systems for the hepatitis C virus.
Bartenschlager R, Lohmann V. Bartenschlager R, et al. Antiviral Res. 2001 Oct;52(1):1-17. doi: 10.1016/s0166-3542(01)00164-4. Antiviral Res. 2001. PMID: 11530183 Review.
Cited by
- Sequential biogenesis of host cell membrane rearrangements induced by hepatitis C virus infection.
Ferraris P, Beaumont E, Uzbekov R, Brand D, Gaillard J, Blanchard E, Roingeard P. Ferraris P, et al. Cell Mol Life Sci. 2013 Apr;70(7):1297-306. doi: 10.1007/s00018-012-1213-0. Epub 2012 Nov 25. Cell Mol Life Sci. 2013. PMID: 23184194 Free PMC article. - Guanylate-binding protein 1 acts as a pro-viral factor for the life cycle of hepatitis C virus.
Bender D, Koulouri A, Wen X, Glitscher M, Schollmeier A, Fernandes da Costa L, Murra RO, Carra GP, Haberger V, Praefcke GJK, Hildt E. Bender D, et al. PLoS Pathog. 2024 Feb 5;20(2):e1011976. doi: 10.1371/journal.ppat.1011976. eCollection 2024 Feb. PLoS Pathog. 2024. PMID: 38315728 Free PMC article. - ARF1 and GBF1 generate a PI4P-enriched environment supportive of hepatitis C virus replication.
Zhang L, Hong Z, Lin W, Shao RX, Goto K, Hsu VW, Chung RT. Zhang L, et al. PLoS One. 2012;7(2):e32135. doi: 10.1371/journal.pone.0032135. Epub 2012 Feb 16. PLoS One. 2012. PMID: 22359663 Free PMC article. - Hepatitis C virus replication is modulated by the interaction of nonstructural protein NS5B and fatty acid synthase.
Huang JT, Tseng CP, Liao MH, Lu SC, Yeh WZ, Sakamoto N, Chen CM, Cheng JC. Huang JT, et al. J Virol. 2013 May;87(9):4994-5004. doi: 10.1128/JVI.02526-12. Epub 2013 Feb 20. J Virol. 2013. PMID: 23427160 Free PMC article. - Effect of Quercetin on Hepatitis C Virus Life Cycle: From Viral to Host Targets.
Rojas Á, Del Campo JA, Clement S, Lemasson M, García-Valdecasas M, Gil-Gómez A, Ranchal I, Bartosch B, Bautista JD, Rosenberg AR, Negro F, Romero-Gómez M. Rojas Á, et al. Sci Rep. 2016 Aug 22;6:31777. doi: 10.1038/srep31777. Sci Rep. 2016. PMID: 27546480 Free PMC article.
References
- Choo QL, et al. Isolation of a cDNA clone derived from a blood-borne non-A non-B viral hepatitis genome. Science. 1989;244:359–362. - PubMed
- Kiyosawa K, et al. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology. 1990;12:671–675. - PubMed
- Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J Gen Virol. 2000;81:1631–1648. - PubMed
- Lohmann V, et al. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- N01 HB027091/HL/NHLBI NIH HHS/United States
- Z01 DK054504/ImNIH/Intramural NIH HHS/United States
- Z99 DK999999/ImNIH/Intramural NIH HHS/United States
- N01-HB-27091/HB/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources