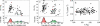Precise temporal modulation in the response of the SOS DNA repair network in individual bacteria - PubMed (original) (raw)
Precise temporal modulation in the response of the SOS DNA repair network in individual bacteria
Nir Friedman et al. PLoS Biol. 2005 Jul.
Abstract
The SOS genetic network is responsible for the repair/bypass of DNA damage in bacterial cells. While the initial stages of the response have been well characterized, less is known about the dynamics of the response after induction and its shutoff. To address this, we followed the response of the SOS network in living individual Escherichia coli cells. The promoter activity (PA) of SOS genes was monitored using fluorescent protein-promoter fusions, with high temporal resolution, after ultraviolet irradiation activation. We find a temporal pattern of discrete activity peaks masked in studies of cell populations. The number of peaks increases, while their amplitude reaches saturation, as the damage level is increased. Peak timing is highly precise from cell to cell and is independent of the stage in the cell cycle at the time of damage. Evidence is presented for the involvement of the umuDC operon in maintaining the pattern of PA and its temporal precision, providing further evidence for the role UmuD cleavage plays in effecting a timed pause during the SOS response, as previously proposed. The modulations in PA we observe share many features in common with the oscillatory behavior recently observed in a mammalian DNA damage response. Our results, which reveal a hitherto unknown modulation of the SOS response, underscore the importance of carrying out dynamic measurements at the level of individual living cells in order to unravel how a natural genetic network operates at the systems level.
Figures
Figure 1. Dynamics of the SOS Response Observed in Individual Cells
(A and B) Snapshots from a time-lapse movie monitoring the fluorescence of live AB1157 E. coli cells taken (A) 8 and (B) 70 min after irradiation with a UV dose of 10 J/m2. Cells are expressing GFP as a reporter for the recA PA [10]. Some cells grow and undergo cell division (e.g., cell #1), while others exhibit filamentation (e.g., cell #2), as a consequence of DNA damage. The exposure time corresponding to (A) is ten times that for (B). (C) Total GFP produced from the recA promoter as a function of time, measured in an individual cell irradiated at 20 J/m2. The full line corresponds to the data after filtration, which is used to compute the PA. (D) PA/PA0 as a function of time for the same cell as in (C). (E–G) Normalized recA promoter activity PA/PA0 as a function of time for cells irradiated at 20, 10, and 50 J/m2, respectively. (H) Average recA PA over all 23 cells in an experiment at 20 J/m2. (I) recA PA/PA0 for two noninducible LexA(Ind−) cells (empty circles), and for two isogenic LexA+ cells (full circles), all irradiated at 20 J/m2. (J) recA PA/PA0 from an unirradiated cell (empty circles), and lacZ PA/PA0 from two cells irradiated at 20 J/m2 (full circles).
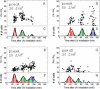
Figure 2. Quantitative Analysis of the Oscillatory Behavior: Distributions of Peaks' Amplitude and Time
Normalized amplitudes of the peaks in recA PA are plotted as function of peak time for individual cells irradiated with a UV dose of (A) 20 J/m2, (B) 50 J/m2. Each point corresponds to one peak in an individual cell. The three clusters in (A) (total of 51 cells) are centered at T 1 = 29 ± 3 min, T 2 = 57 ± 5 min, and T 3 = 93 ± 4 min (mean ± standard deviation). The average normalized promoter activities corresponding to these three clusters are: PA1 = 15 ± 4, PA2 = 19 ± 5, and PA3 = 13 ± 4 in units of PA0. (C and D) Histograms of peak times corresponding to (A) and (B), ranking each peak by its order of appearance: red: first peak in a trace of PA(t) of an individual bacteria; green: a second peak in its trace; blue: a third peak in its trace. Black lines: fits to the histograms with a sum of Gaussians. (E) Peak lexA PA as function of peak time for individual cells and (F) its corresponding histogram. (G) Peak umuDC PA as function of peak time for individual cells and (H) its corresponding histogram. Cells in the experiments (E) and (G) were irradiated with a UV dose of 20 J/m2.
Figure 3. Correlations between the Peaks' Time and Bacterial Growth Parameters
(A) Scatter plot of the time of the first peak, T 1, (normalized by the population average value <_T_ 1>) vs. the length of the cell at the time of UV irradiation, L 0 (normalized by the population average <_L_ 0>). All cells from all UV doses are included; each point represents an individual cell. No correlation between the two quantities is observed (correlation coefficient = 0.02, p = 0.82). (B) Scatter plot of 1/T 1 as a function of the growth rate, 1/TD, of individual cells irrespective of dose. A significant correlation is observed (correlation coefficient = 0.66, p < 10–4). A linear fit to the data yields a slope of 1.0 ± 0.1. Inset: Cell doubling time _TD_ grows monotonically as a function of UV dose. (C) Time of the second (_T_ 2) and third (_T_ 3) peaks is plotted against the time of appearance of the first peak (_T_ 1). Each point corresponds to an individual cell. The data for _T_ 2 correspond to 10 J/m2 (green), 20 J/m2 (red), 35 J/m2 (blue), and 50 J/m2 (magenta). The data for _T_ 3 are shown in black irrespective of radiation dose for the sake of clarity. Full lines, _T_ 2 = 2_T_ 1, and _T_ 3 = 3_T_ 1 are shown as a guide to the eye. Averaging over all experiments at all UV doses we obtain: <_T_ 2/_T_ 1> = 1.99 ± 0.02 (mean ± standard error, over 132 cells), whereas <_T_ 3/_T_ 1> = 2.99 ± 0.06 (over the 60 cells that show a third peak). Peak times in A, B, and C are from measurements performed with the recA promoter.
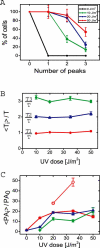
Figure 4. Dependence of Response Parameters on UV Dose
(A) Percentage of cells exhibiting at least zero, one, two, or three peaks at different damage levels. (B) Dependence of the mean peak time <_Ti_>, normalized by T = (1/TD + 1/τ)–1 (τ = 68 min), on UV dose. (C) Dependence of the mean normalized peak height <PA_i_>/PA0 on UV dose. Wild type (full symbols), ΔumuDC (empty symbols). Same color and symbol convention as in (B): <PA1>/PA0 (red circles), <PA2>/PA0 (blue triangles), and <PA3>/PA0 (green diamonds). In wild-type cells, the amplitude of peaks saturates at approximately 20 PA0 for UV doses greater than 20 J/m2. ΔumuDC cells do not show this saturation and reach much higher PA levels. Parameters in (A), (B), and (C) are from measurements performed with the recA promoter.
Figure 5. Effects of the umuDC Operon on the Temporal Modulation of Promoter Activity
(A) Peak PA of the recA promoter as a function of peak time for individual ΔumuDC cells and (B) its corresponding histogram of the peak times. The amplitude and timing of the first peak are more correlated (correlation coefficient = 0.37, p = 0.002) than in wild-type cells (Figure 2A, correlation coefficient = 0.18, p = 0.13). (C) Peak PA of the recA promoter as a function of peak time for individual AB1157 cells transformed with a noncleavable umuD mutant gene (K97A) expressed from a plasmid, and (D) its corresponding histogram of the peak times. The experiments were carried out at 20 J/m2. (E) Scatter plot of 1/T 1 as a function of the growth rate, 1/TD, of individual ΔumuDC cells irrespective of UV dose. In contrast with wild-type behavior (see Figure 3B), experiments with ΔumuDC mutants show that the timing of first peak maxima and cell growth rate 1/TD are poorly correlated (correlation coefficient = 0.15, p = 0.06, compared with a correlation coefficient = 0.66, p < 10–4 measured for AB1157 cells).
Comment in
- After 30 years of study, the bacterial SOS response still surprises us.
Michel B. Michel B. PLoS Biol. 2005 Jul;3(7):e255. doi: 10.1371/journal.pbio.0030255. Epub 2005 Jul 12. PLoS Biol. 2005. PMID: 16000023 Free PMC article.
Similar articles
- The SOS response and induced mutagenesis.
Battista JR, Donnelly CE, Ohta T, Walker GC. Battista JR, et al. Prog Clin Biol Res. 1990;340A:169-78. Prog Clin Biol Res. 1990. PMID: 2167481 Review. No abstract available. - Alleviation of EcoK DNA restriction in Escherichia coli and involvement of umuDC activity.
Hiom KJ, Sedgwick SG. Hiom KJ, et al. Mol Gen Genet. 1992 Jan;231(2):265-75. doi: 10.1007/BF00279800. Mol Gen Genet. 1992. PMID: 1310522 - A role for the umuDC gene products of Escherichia coli in increasing resistance to DNA damage in stationary phase by inhibiting the transition to exponential growth.
Murli S, Opperman T, Smith BT, Walker GC. Murli S, et al. J Bacteriol. 2000 Feb;182(4):1127-35. doi: 10.1128/JB.182.4.1127-1135.2000. J Bacteriol. 2000. PMID: 10648540 Free PMC article. - Survival and SOS response induction in ultraviolet B irradiated Escherichia coli cells with defective repair mechanisms.
Prada Medina CA, Aristizabal Tessmer ET, Quintero Ruiz N, Serment-Guerrero J, Fuentes JL. Prada Medina CA, et al. Int J Radiat Biol. 2016 Jun;92(6):321-8. doi: 10.3109/09553002.2016.1152412. Epub 2016 Mar 11. Int J Radiat Biol. 2016. PMID: 26967458 - The "tale" of UmuD and its role in SOS mutagenesis.
Gonzalez M, Woodgate R. Gonzalez M, et al. Bioessays. 2002 Feb;24(2):141-8. doi: 10.1002/bies.10040. Bioessays. 2002. PMID: 11835278 Review.
Cited by
- Inherent regulatory asymmetry emanating from network architecture in a prevalent autoregulatory motif.
Ali MZ, Parisutham V, Choubey S, Brewster RC. Ali MZ, et al. Elife. 2020 Aug 18;9:e56517. doi: 10.7554/eLife.56517. Elife. 2020. PMID: 32808926 Free PMC article. - Mutation as a stress response and the regulation of evolvability.
Galhardo RS, Hastings PJ, Rosenberg SM. Galhardo RS, et al. Crit Rev Biochem Mol Biol. 2007 Sep-Oct;42(5):399-435. doi: 10.1080/10409230701648502. Crit Rev Biochem Mol Biol. 2007. PMID: 17917874 Free PMC article. Review. - Using movies to analyse gene circuit dynamics in single cells.
Locke JC, Elowitz MB. Locke JC, et al. Nat Rev Microbiol. 2009 May;7(5):383-92. doi: 10.1038/nrmicro2056. Nat Rev Microbiol. 2009. PMID: 19369953 Free PMC article. Review. - P1 Ref Endonuclease: A Molecular Mechanism for Phage-Enhanced Antibiotic Lethality.
Ronayne EA, Wan YC, Boudreau BA, Landick R, Cox MM. Ronayne EA, et al. PLoS Genet. 2016 Jan 14;12(1):e1005797. doi: 10.1371/journal.pgen.1005797. eCollection 2016 Jan. PLoS Genet. 2016. PMID: 26765929 Free PMC article. - Genomic survey and expression analysis of DNA repair genes in the genus Leptospira.
Martins-Pinheiro M, Schons-Fonseca L, da Silva JB, Domingos RH, Momo LH, Simões AC, Ho PL, da Costa RM. Martins-Pinheiro M, et al. Mol Genet Genomics. 2016 Apr;291(2):703-22. doi: 10.1007/s00438-015-1135-2. Epub 2015 Nov 2. Mol Genet Genomics. 2016. PMID: 26527082
References
- Little JW. The SOS regulatory system. In: Lin ECC, Simon A, editors. Regulation of gene expression in Escherichia coli. Austin (Texas): Landes; 1996. pp. 453–479.
- Friedberg EC, Walker GC, Siede W. DNA repair and mutagenesis. Washington (DC): ASM Press; 1995. 698 pp.
- Fernandez de Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ. Identification of additional genes belonging to the LexA regulon in Escherichia coli . Mol Microbiol. 2000;35:1560–1572. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources