A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts - PubMed (original) (raw)
Review
A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts
Michael Y Galperin. BMC Microbiol. 2005.
Abstract
Background: Analysis of complete microbial genomes showed that intracellular parasites and other microorganisms that inhabit stable ecological niches encode relatively primitive signaling systems, whereas environmental microorganisms typically have sophisticated systems of environmental sensing and signal transduction.
Results: This paper presents results of a comprehensive census of signal transduction proteins--histidine kinases, methyl-accepting chemotaxis receptors, Ser/Thr/Tyr protein kinases, adenylate and diguanylate cyclases and c-di-GMP phosphodiesterases--encoded in 167 bacterial and archaeal genomes, sequenced by the end of 2004. The data have been manually checked to avoid false-negative and false-positive hits that commonly arise during large-scale automated analyses and compared against other available resources. The census data show uneven distribution of most signaling proteins among bacterial and archaeal phyla. The total number of signal transduction proteins grows approximately as a square of genome size. While histidine kinases are found in representatives of all phyla and are distributed according to the power law, other signal transducers are abundant in certain phylogenetic groups but virtually absent in others.
Conclusion: The complexity of signaling systems differs even among closely related organisms. Still, it usually can be correlated with the phylogenetic position of the organism, its lifestyle, and typical environmental challenges it encounters. The number of encoded signal transducers (or their fraction in the total protein set) can be used as a measure of the organism's ability to adapt to diverse conditions, the 'bacterial IQ', while the ratio of transmembrane receptors to intracellular sensors can be used to define whether the organism is an 'extrovert', actively sensing the environmental parameters, or an 'introvert', more concerned about its internal homeostasis. Some of the microorganisms with the highest IQ, including the current leader Wolinella succinogenes, are found among the poorly studied beta-, delta- and epsilon-proteobacteria. Among all bacterial phyla, only cyanobacteria appear to be true introverts, probably due to their capacity to conduct oxygenic photosynthesis, using a complex system of intracellular membranes. The census data, available at http://www.ncbi.nlm.nih.gov/Complete\_Genomes/SignalCensus.html, can be used to get an insight into metabolic and behavioral propensities of each given organism and improve prediction of the organism's properties based solely on its genome sequence.
Figures
Figure 1
Growth in the number of signal transduction proteins encoded in complete microbial genomes. A. Histidine kinases (circles), methyl-accepting chemotaxis proteins (MCPs, squares) and Ser/Thr/Tyr protein kinases (STYKs, triangles). B. Diguanylate cyclases (GGDEF domains, diamonds), c-di-GMP-specific phosphodiesterases (EAL domains, triangles), and adenylate cyclases (circles). Open symbols indicate the total number of proteins with the corresponding domains, closed symbols – the number of membrane-bound proteins of each kind.
Figure 2
The total number of encoded signal transducers proteins grows with genome size. A. Distribution of signal transduction proteins among free-living bacteria and archaea (squares) and obligate pathogens (closed squares). Organisms with other status (symbionts and other commensals) are indicated by triangles. The best-fit line represents data from all species. B. Distribution of signal transduction proteins among organisms of different phylogenetic lineages. The symbols indicate members of the following phyla: Actinobacteria, black circles; Cyanobacteria, open circles (light blue); Alpha-proteobacteria, closed diamonds (dark brown); Beta-, Delta-, and Epsilon-proteobacteria, open diamonds (yellow); Gamma-proteobacteria, closed squares (dark blue); Firmicutes, open squares (magenta); members of other bacterial phyla (Aquificales, Bacteroidetes, Chlamydiae, Deinococcus-Thermus, Planctomycetes, Spirochetes, Thermotoga), closed triangles (red); Archaea, open triangles (yellow).
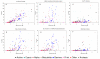
Figure 3
Phylogenetic distribution of certain types of signal transducers. A. Histidine kinases. B. Methyl-accepting chemotaxis proteins. C. Ser/Thr/Tyr-protein kinases. D. GGDEF domains (active and inactive diguanylate cyclases). E. EAL domains (active and inactive c-di-GMP phosphodiesterases). F. Adenylate cyclases. The symbols for various phyla are shown at the bottom and are the same as in Fig. 2b.
Figure 4
Distribution of the signal transduction proteins follows the power law. A. Distribution of histidine kinases. B. Distribution of the total number of all signal transduction proteins except for histidine kinases encoded in a given genome. The symbols for various phyla are as in Fig. 2b.
Figure 5
Phylogenetic distribution of membrane-bound signal transduction proteins. A. Phylogenetic distribution of the total number of transmembrane signal transducers. The best-fit lines are shown for proteins from gamma-bacteria (dark blue) and cyanobacteria (cyan). The symbols for various phyla are as in Fig. 2b. B. Transmembrane histidine kinases (squares), Ser/Thr kinases (circles) and diguanylate cyclases (triangles) in all proteobacteria (open symbols) and cyanobacteria (closed symbols). The best-fit lines are shown for proteobacterial (dark blue) and cyanobacterial (cyan) histidine kinases. C. Phylogenetic distribution of transmembrane histidine kinases. The best-fit lines are shown for actinobacterial (black) and cyanobacterial (cyan) histidine kinases.
Similar articles
- Phyletic Distribution and Lineage-Specific Domain Architectures of Archaeal Two-Component Signal Transduction Systems.
Galperin MY, Makarova KS, Wolf YI, Koonin EV. Galperin MY, et al. J Bacteriol. 2018 Mar 12;200(7):e00681-17. doi: 10.1128/JB.00681-17. Print 2018 Apr 1. J Bacteriol. 2018. PMID: 29263101 Free PMC article. - Interplay of heritage and habitat in the distribution of bacterial signal transduction systems.
Galperin MY, Higdon R, Kolker E. Galperin MY, et al. Mol Biosyst. 2010 Apr;6(4):721-8. doi: 10.1039/b908047c. Epub 2010 Feb 9. Mol Biosyst. 2010. PMID: 20237650 Free PMC article. - Sentra: a database of signal transduction proteins for comparative genome analysis.
D'Souza M, Glass EM, Syed MH, Zhang Y, Rodriguez A, Maltsev N, Galperin MY. D'Souza M, et al. Nucleic Acids Res. 2007 Jan;35(Database issue):D271-3. doi: 10.1093/nar/gkl949. Epub 2006 Nov 29. Nucleic Acids Res. 2007. PMID: 17135204 Free PMC article. - Identification of sensory and signal-transducing domains in two-component signaling systems.
Galperin MY, Nikolskaya AN. Galperin MY, et al. Methods Enzymol. 2007;422:47-74. doi: 10.1016/S0076-6879(06)22003-2. Methods Enzymol. 2007. PMID: 17628134 Free PMC article. Review. - What bacteria want.
Galperin MY. Galperin MY. Environ Microbiol. 2018 Dec;20(12):4221-4229. doi: 10.1111/1462-2920.14398. Epub 2018 Oct 25. Environ Microbiol. 2018. PMID: 30187651 Free PMC article. Review.
Cited by
- The cognitive cell: bacterial behavior reconsidered.
Lyon P. Lyon P. Front Microbiol. 2015 Apr 14;6:264. doi: 10.3389/fmicb.2015.00264. eCollection 2015. Front Microbiol. 2015. PMID: 25926819 Free PMC article. Review. - Evolution of two-component signal transduction systems.
Capra EJ, Laub MT. Capra EJ, et al. Annu Rev Microbiol. 2012;66:325-47. doi: 10.1146/annurev-micro-092611-150039. Epub 2012 Jun 28. Annu Rev Microbiol. 2012. PMID: 22746333 Free PMC article. Review. - Trigger phosphodiesterases as a novel class of c-di-GMP effector proteins.
Hengge R. Hengge R. Philos Trans R Soc Lond B Biol Sci. 2016 Nov 5;371(1707):20150498. doi: 10.1098/rstb.2015.0498. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27672149 Free PMC article. Review. - The role of two-component regulation systems in the physiology of the bacterial cell.
Bekker M, Teixeira de Mattos MJ, Hellingwerf KJ. Bekker M, et al. Sci Prog. 2006;89(Pt 3-4):213-42. doi: 10.3184/003685006783238308. Sci Prog. 2006. PMID: 17338439 Free PMC article. Review. - The evolution of two-component systems in bacteria reveals different strategies for niche adaptation.
Alm E, Huang K, Arkin A. Alm E, et al. PLoS Comput Biol. 2006 Nov 3;2(11):e143. doi: 10.1371/journal.pcbi.0020143. PLoS Comput Biol. 2006. PMID: 17083272 Free PMC article.
References
- Sondej M, Weinglass AB, Peterkofsky A, Kaback HR. Binding of enzyme IIAGlc, a component of the phosphoenolpyruvate:sugar phosphotransferase system, to the Escherichia coli lactose permease. Biochemistry. 2002;41:5556–5565. - PubMed
- Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett. 2001;203:11–21. - PubMed
- Mizuno T, Kaneko T, Tabata S. Compilation of all genes encoding bacterial two-component signal transducers in the genome of the cyanobacterium, Synechocystis sp. strain PCC 6803. DNA Res. 1996;3:407–414. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources