Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression - PubMed (original) (raw)
Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression
María Eugenia Zanetti et al. Plant Physiol. 2005 Jun.
Abstract
Immunoaffinity purification of polyribosomes (polysomes) from crude leaf extracts of Arabidopsis (Arabidopsis thaliana) was achieved with transgenic genotypes that overexpress a translational fusion of a ribosomal protein (RP) with a His(6)-FLAG dual epitope tag. In plants with a cauliflower mosaic virus 35S:HF-RPL18 transgene immunopurification with anti-FLAG agarose beads yielded 60-Svedberg ribosomal subunits, intact 80-Svedberg monosomes and polysomes. Sucrose density gradient fractionation of the purified complexes demonstrated that the distribution of polysome size was similar in crude cell extracts and the purified complexes. The immunopurified complexes included putative cytosolic RPs of Arabidopsis and ribosome-associated proteins, as well as full-length transcripts of high and low abundance. Whole-genome profiling using long DNA oligonucleotide-based microarrays provided a high level of reproducibility between polysomal mRNA samples immunopurified from two independent biological replicates (r approximately 0.90). Comparison of immunopurified and total cellular RNA samples revealed that for most of the genes, the mRNAs were associated with the epitope-tagged polysomal complexes, with an average relative level of association of 62.06% +/- 4.39%. The results demonstrate that the immunopurification of polysomes can be a valuable tool for the quantification of mRNAs present in translation complexes in plant cells. This technology can be extended to evaluation of mRNA populations at the cell- or tissue-specific level by regulation of the tagged RP with distinct promoters.
Figures
Figure 1.
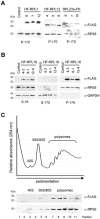
Epitope-tagged RPs are associated with translating ribosomes. A, Ribosomes were pelleted by centrifugation from detergent-treated cell extracts of control (transformed with empty HF T-DNA vector) and transgenic lines of Arabidopsis. Proteins (30 _μ_g per lane) were separated by 15% (w/v) SDS-PAGE, transferred onto nitrocellulose membranes, and the epitope-tagged RPs were immunodetected with the anti-FLAG-horseradish-peroxidase-conjugated monoclonal antibody (_α_-FLAG). Identical membranes were used for immunodetection of RPS6, a component of the small ribosomal subunit (_α_-RPS6). The migration of molecular mass markers is indicated on the left. B, Crude extracts (S-16), postribosomal supernatants (S-170), and ribosome pellet fractions (P-170) were prepared from control and 35S:HF-RPL18 Arabidopsis lines and analyzed as described for A with _α_-FLAG, _α_-RPS6, and an antiserum against a soluble cytosolic GAPDH (_α_-GAPDH). C, The ribosomal pellet fraction (P-170) from 35S:HF-RPL18 plants was fractionated by ultracentrifugation through 20% to 60% (w/v) Suc density gradients, and the UV absorbance (254 nm) profile was recorded. The 11 gradient fractions were analyzed by immunoblot analysis with _α_-FLAG and _α_-RPS6. The position of the ribosomal subunits (40S, 60S), monosomes (80S), and polysomes are indicated.
Figure 2.
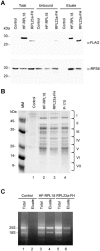
Immunopurification of Arabidopsis 80S ribosomes from crude leaf extracts by use of epitope-tagged RPL18 and RPL23a. Leaf proteins from control (transformed with the empty HF T-DNA vector), 35S:HF-RPL18, or 35S:RPL23a-FH transgenic lines were extracted in PEB, and approximately 300 OD260 units from each sample were incubated with anti-FLAG agarose beads for isolation of protein complexes that contained the HF epitope-tagged RP. A, Total, unbound, or immunopurified (Eluate) proteins were separated by 15% (w/v) SDS-PAGE, transferred onto a nitrocellulose membrane, and immunodetected with _α_-FLAG and _α_-RPS6 as described in Figure 1. Molecular mass markers are indicated on the left. B, Proteins (30 _μ_g per lane) that were coaffinity-purified with HF-RPL18 (lane 2) or with RPL23a-FH (lane 3) and RPs prepared by conventional ultracentrifugation (P-170, lane 4) were visualized by Coomassie Blue staining. Lane 1 represents a proportional volume of the sample immunopurified from control plants. Proteins copurified with HF-RPL18 (lane 2) were identified by mass spectrometric analyses (see also Supplemental Table I). The position of the seven gel sections analyzed by MS is indicated on the right. C, RNA was extracted from leaf crude extracts (Total) or the immunopurified material (Eluate) of control, 35S:HF-RPL18, and 35S:RPL23a-FH transgenic lines by guanidine-HCl/ethanol precipitation. Two micrograms of total RNA from control, 35S:HF-RPL18, and 35S:RPL23a-FH lines (lanes 1, 3, and 5) and eluted RNA from 35S:HF-RPL18 and 35S:RPL23a-FH lines (lanes 4 and 6) were resolved in a 1.2% (w/v) agarose gel containing 10 _μ_g mL−1 of ethidium bromide. Lane 2 is a proportional volume of the eluted sample from control plants. The 25S and 18S rRNAs are indicated.
Figure 3.
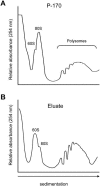
Sedimentation characteristics of ultracentrifugation-concentrated and immunopurified ribosomes on Suc density gradients. Leaf ribosomes were isolated by ultracentrifugation through a 1.6
m
Suc cushion or by affinity purification with anti-FLAG agarose beads. Ribosomes (approximately 15 OD260 units) were fractionated by ultracentrifugation through 20% to 60% (w/v) Suc density gradients, and the UV absorbance (254 nm) profile was recorded. A, Absorbance profile of ribosomes purified by conventional ultracentrifugation (P-170). B, Absorbance profile of ribosomes copurified with HF-RPL18 (Eluate). The positions of the 60S ribosomal subunit, 80S monosomes, and polysomes are indicated.
Figure 4.
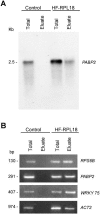
Analysis of mRNAs associated with immunopurified ribosomes. A, Approximately 8 _μ_g of total or immunopurified (Eluate) RNA was resolved in a formaldehyde agarose gel and transferred onto a nylon membrane. In the control sample a volume proportional to HF-RPL18 eluate sample was loaded. The membrane was hybridized to [32P]-labeled PABP2 and exposed to x-ray film. The size (kilobase) of the PABP2 transcript is indicated on the left. B, Total or immunopurified (Eluate) RNA (50 ng) isolated from control and 35S:HF-RPL18 transgenic lines were subjected to one-step RT-PCR amplification with gene-specific primers for RPS6, PABP, WRKY75, and ACT2. Twenty-five cycles of amplification were used for RPS6, PABP, and ACT2, and 30 cycles were used for WRKY75. The PCR products were separated in a 2% (w/v) agarose gel containing 10 _μ_g mL−1 of ethidium bromide. The size (basepair) of each product is indicated.
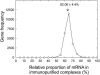
Figure 5.
Frequency distribution of mRNA levels in polysomal complexes. The relative percentage of each mRNA found in the immunopurified polysomal sample was determined for 23,471 genes as described in “Material and Methods,” and used to bin the number of expressed genes (frequency) at 5% intervals. The average value and
sd
of percentage of association with immunopurified polysomes (62.06% ± 4.39%) is indicated.
Similar articles
- Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods.
Mustroph A, Juntawong P, Bailey-Serres J. Mustroph A, et al. Methods Mol Biol. 2009;553:109-26. doi: 10.1007/978-1-60327-563-7_6. Methods Mol Biol. 2009. PMID: 19588103 Review. - Cold shock protein 1 chaperones mRNAs during translation in Arabidopsis thaliana.
Juntawong P, Sorenson R, Bailey-Serres J. Juntawong P, et al. Plant J. 2013 Jun;74(6):1016-28. doi: 10.1111/tpj.12187. Epub 2013 May 6. Plant J. 2013. PMID: 23551487 - Detection of Argonaute 1 Association with Polysomes in Arabidopsis thaliana.
Lecampion C, Lanet E, Robaglia C. Lecampion C, et al. Methods Mol Biol. 2017;1640:137-143. doi: 10.1007/978-1-4939-7165-7_9. Methods Mol Biol. 2017. PMID: 28608339 - Genome-wide analysis of transcript abundance and translation in Arabidopsis seedlings subjected to oxygen deprivation.
Branco-Price C, Kawaguchi R, Ferreira RB, Bailey-Serres J. Branco-Price C, et al. Ann Bot. 2005 Sep;96(4):647-60. doi: 10.1093/aob/mci217. Epub 2005 Aug 4. Ann Bot. 2005. PMID: 16081496 Free PMC article. - Purification, identification, and functional analysis of polysomes from the human pathogen Staphylococcus aureus.
Brielle R, Pinel-Marie ML, Chat S, Gillet R, Felden B. Brielle R, et al. Methods. 2017 Mar 15;117:59-66. doi: 10.1016/j.ymeth.2016.10.003. Epub 2016 Oct 8. Methods. 2017. PMID: 27729294 Review.
Cited by
- Legume-rhizobia symbiosis: Translatome analysis.
Sainz MM, Sotelo-Silveira M, Filippi CV, Zardo S. Sainz MM, et al. Genet Mol Biol. 2024 Jul 1;47Suppl 1(Suppl 1):e20230284. doi: 10.1590/1678-4685-GMB-2023-0284. eCollection 2024. Genet Mol Biol. 2024. PMID: 38954532 Free PMC article. - Florigen-producing cells express FPF1-LIKE PROTEIN 1 that accelerates flowering and stem growth in long days with sunlight red/far-red ratio in Arabidopsis.
Takagi H, Lee N, Hempton AK, Purushwani S, Notaguchi M, Yamauchi K, Shirai K, Kawakatsu Y, Uehara S, Albers WG, Downing BLR, Ito S, Suzuki T, Matsuura T, Mori IC, Mitsuda N, Kurihara D, Matsushita T, Song YH, Sato Y, Nomoto M, Tada Y, Hanada K, Cuperus JT, Queitsch C, Imaizumi T. Takagi H, et al. bioRxiv [Preprint]. 2024 Apr 29:2024.04.26.591289. doi: 10.1101/2024.04.26.591289. bioRxiv. 2024. PMID: 38746097 Free PMC article. Preprint. - Isolation of Cytosolic Ribosomes Associated with Plant Mitochondria and Chloroplasts.
Dimnet L, Salinas-Giegé T, Pullara S, Moyet L, Genevey C, Kuntz M, Duchêne AM, Rolland N. Dimnet L, et al. Methods Mol Biol. 2024;2776:289-302. doi: 10.1007/978-1-0716-3726-5_18. Methods Mol Biol. 2024. PMID: 38502512 - Plant mRNAs move into a fungal pathogen via extracellular vesicles to reduce infection.
Wang S, He B, Wu H, Cai Q, Ramírez-Sánchez O, Abreu-Goodger C, Birch PRJ, Jin H. Wang S, et al. Cell Host Microbe. 2024 Jan 10;32(1):93-105.e6. doi: 10.1016/j.chom.2023.11.020. Epub 2023 Dec 15. Cell Host Microbe. 2024. PMID: 38103543 - Light-induced LLPS of the CRY2/SPA1/FIO1 complex regulating mRNA methylation and chlorophyll homeostasis in Arabidopsis.
Jiang B, Zhong Z, Gu L, Zhang X, Wei J, Ye C, Lin G, Qu G, Xiang X, Wen C, Hummel M, Bailey-Serres J, Wang Q, He C, Wang X, Lin C. Jiang B, et al. Nat Plants. 2023 Dec;9(12):2042-2058. doi: 10.1038/s41477-023-01580-0. Epub 2023 Dec 8. Nat Plants. 2023. PMID: 38066290 Free PMC article.
References
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920 - PubMed
- Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E (2001) Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol Cell 8: 517–529 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous