Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques - PubMed (original) (raw)
. 2005 Jun 21;102(25):9014-9.
doi: 10.1073/pnas.0503671102. Epub 2005 Jun 14.
Stephen F Porcella, Morag R Graham, Robin M Ireland, Claire A Johnson, Stacy M Ricklefs, Imran Babar, Larye D Parkins, Romina A Romero, G Judson Corn, Don J Gardner, John R Bailey, Michael J Parnell, James M Musser
Affiliations
- PMID: 15956184
- PMCID: PMC1150296
- DOI: 10.1073/pnas.0503671102
Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques
Kimmo Virtaneva et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Identification of the genetic events that contribute to host-pathogen interactions is important for understanding the natural history of infectious diseases and developing therapeutics. Transcriptome studies conducted on pathogens have been central to this goal in recent years. However, most of these investigations have focused on specific end points or disease phases, rather than analysis of the entire time course of infection. To gain a more complete understanding of how bacterial gene expression changes over time in a primate host, the transcriptome of group A Streptococcus (GAS) was analyzed during an 86-day infection protocol in 20 cynomolgus macaques with experimental pharyngitis. The study used 260 custom Affymetrix (Santa Clara, CA) chips, and data were confirmed by TaqMan analysis. Colonization, acute, and asymptomatic phases of disease were identified. Successful colonization and severe inflammation were significantly correlated with an early onset of superantigen gene expression. The differential expression of two-component regulators covR and spy0680 (M1_spy0874) was significantly associated with GAS colony-forming units, inflammation, and phases of disease. Prophage virulence gene expression and prophage induction occurred predominantly during high pathogen cell densities and acute inflammation. We discovered that temporal changes in the GAS transcriptome were integrally linked to the phase of clinical disease and host-defense response. Knowledge of the gene expression patterns characterizing each phase of pathogen-host interaction provides avenues for targeted investigation of proven and putative virulence factors and genes of unknown function and will assist vaccine research.
Figures
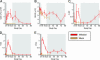
Fig. 1.
Five medical parameters used in expression analysis of GAS PHG. Each parameter is depicted by two-way ANOVA plot, with the measurement on the y axis and the sample collection day of the study on x axis. The infection phase (days 0–86) is marked with a red line, and the mock phase (days 0–32) is labeled brown. SEM is indicated by error bars. (A) GAS colony count as measured from tonsillar throat swabs (CFU). (B) Percentage of polymorphonuclear leukocytes in peripheral venous blood (SEGS). (C) C-RP levels in blood (mg/liter; C-RP). (D) TON score on a scale of 1–4. (E) PHG severity score on a scale of 0–3 (PHG). Colonization and asymptomatic carriage phases are indicated by gray shading.
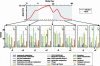
Fig. 2.
Functional category analysis of GAS gene expression data. GAS CFU were plotted on the y axis, and the study day was plotted on the x axis. The infection phase (days 0–86) is indicated by a red line, and the mock phase (days 0–32) is labeled in brown. Number of GAS genes belonging to 17 functional categories were plotted based on their correlation with GAS CFU. Genes having positive (+Z) or negative (–Z) expression correlation with GAS CFU are shown by histograms at three phases. Colonization and asymptomatic carriage phases are indicated by gray shading.
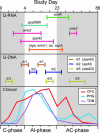
Fig. 3.
TaqMan RNA quantitative PCR (Q-PCR) confirmation of expression of GAS SAgs involved in inflammation (smeZ, speA2, and speJ), complement resistance regulon (mga, emm1, sic, and scpA), and TCS regulation (covR and spy0680/M1_ spy0874). Induction of strain MGAS5005 prophage (Φ1, Φ2, and Φ3) as confirmed by TaqMan DNA Q-PCR. Bars indicate time points at which the level of RNA or DNA is at least 1.25-fold or greater than mean expression level. Clinical parameters for GAS CFU (red), PHG (blue), and TON (purple) are shown. Colonization and asymptomatic carriage phases are indicated by gray shading.
Similar articles
- Group A Streptococcus gene expression in humans and cynomolgus macaques with acute pharyngitis.
Virtaneva K, Graham MR, Porcella SF, Hoe NP, Su H, Graviss EA, Gardner TJ, Allison JE, Lemon WJ, Bailey JR, Parnell MJ, Musser JM. Virtaneva K, et al. Infect Immun. 2003 Apr;71(4):2199-207. doi: 10.1128/IAI.71.4.2199-2207.2003. Infect Immun. 2003. PMID: 12654842 Free PMC article. - Interactome analysis of longitudinal pharyngeal infection of cynomolgus macaques by group A Streptococcus.
Shea PR, Virtaneva K, Kupko JJ 3rd, Porcella SF, Barry WT, Wright FA, Kobayashi SD, Carmody A, Ireland RM, Sturdevant DE, Ricklefs SM, Babar I, Johnson CA, Graham MR, Gardner DJ, Bailey JR, Parnell MJ, Deleo FR, Musser JM. Shea PR, et al. Proc Natl Acad Sci U S A. 2010 Mar 9;107(10):4693-8. doi: 10.1073/pnas.0906384107. Epub 2010 Feb 23. Proc Natl Acad Sci U S A. 2010. PMID: 20179180 Free PMC article. - Streptococcus pyogenes genes that promote pharyngitis in primates.
Zhu L, Olsen RJ, Beres SB, Saavedra MO, Kubiak SL, Cantu CC, Jenkins L, Waller AS, Sun Z, Palzkill T, Porter AR, DeLeo FR, Musser JM. Zhu L, et al. JCI Insight. 2020 Jun 4;5(11):e137686. doi: 10.1172/jci.insight.137686. JCI Insight. 2020. PMID: 32493846 Free PMC article. - Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions.
Kreikemeyer B, McIver KS, Podbielski A. Kreikemeyer B, et al. Trends Microbiol. 2003 May;11(5):224-32. doi: 10.1016/s0966-842x(03)00098-2. Trends Microbiol. 2003. PMID: 12781526 Review. - Group A streptococcal pharyngitis: Immune responses involved in bacterial clearance and GAS-associated immunopathologies.
Soderholm AT, Barnett TC, Sweet MJ, Walker MJ. Soderholm AT, et al. J Leukoc Biol. 2018 Feb;103(2):193-213. doi: 10.1189/jlb.4MR0617-227RR. Epub 2017 Dec 29. J Leukoc Biol. 2018. PMID: 28951419 Review.
Cited by
- MalE of group A Streptococcus participates in the rapid transport of maltotriose and longer maltodextrins.
Shelburne SA 3rd, Fang H, Okorafor N, Sumby P, Sitkiewicz I, Keith D, Patel P, Austin C, Graviss EA, Musser JM, Chow DC. Shelburne SA 3rd, et al. J Bacteriol. 2007 Apr;189(7):2610-7. doi: 10.1128/JB.01539-06. Epub 2007 Jan 26. J Bacteriol. 2007. PMID: 17259319 Free PMC article. - Intranasal exposure to bacterial superantigens induces airway inflammation in HLA class II transgenic mice.
Rajagopalan G, Iijima K, Singh M, Kita H, Patel R, David CS. Rajagopalan G, et al. Infect Immun. 2006 Feb;74(2):1284-96. doi: 10.1128/IAI.74.2.1284-1296.2006. Infect Immun. 2006. PMID: 16428778 Free PMC article. - Mga is sufficient to activate transcription in vitro of sof-sfbX and other Mga-regulated virulence genes in the group A Streptococcus.
Almengor AC, Walters MS, McIver KS. Almengor AC, et al. J Bacteriol. 2006 Mar;188(6):2038-47. doi: 10.1128/JB.188.6.2038-2047.2006. J Bacteriol. 2006. PMID: 16513733 Free PMC article. - Microevolution of group A streptococci in vivo: capturing regulatory networks engaged in sociomicrobiology, niche adaptation, and hypervirulence.
Aziz RK, Kansal R, Aronow BJ, Taylor WL, Rowe SL, Kubal M, Chhatwal GS, Walker MJ, Kotb M. Aziz RK, et al. PLoS One. 2010 Apr 14;5(4):e9798. doi: 10.1371/journal.pone.0009798. PLoS One. 2010. PMID: 20418946 Free PMC article. - Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences.
Nasser W, Beres SB, Olsen RJ, Dean MA, Rice KA, Long SW, Kristinsson KG, Gottfredsson M, Vuopio J, Raisanen K, Caugant DA, Steinbakk M, Low DE, McGeer A, Darenberg J, Henriques-Normark B, Van Beneden CA, Hoffmann S, Musser JM. Nasser W, et al. Proc Natl Acad Sci U S A. 2014 Apr 29;111(17):E1768-76. doi: 10.1073/pnas.1403138111. Epub 2014 Apr 14. Proc Natl Acad Sci U S A. 2014. PMID: 24733896 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases