Recruitment of DNA methyltransferase I to DNA repair sites - PubMed (original) (raw)
Recruitment of DNA methyltransferase I to DNA repair sites
Oliver Mortusewicz et al. Proc Natl Acad Sci U S A. 2005.
Abstract
In mammalian cells, the replication of genetic and epigenetic information is directly coupled; however, little is known about the maintenance of epigenetic information in DNA repair. Using a laser microirradiation system to introduce DNA lesions at defined subnuclear sites, we tested whether the major DNA methyltransferase (Dnmt1) or one of the two de novo methyltransferases (Dnmt3a, Dnmt3b) are recruited to sites of DNA repair in vivo. Time lapse microscopy of microirradiated mammalian cells expressing GFP-tagged Dnmt1, Dnmt3a, or Dnmt3b1 together with red fluorescent protein-tagged proliferating cell nuclear antigen (PCNA) revealed that Dnmt1 and PCNA accumulate at DNA damage sites as early as 1 min after irradiation in S and non-S phase cells, whereas recruitment of Dnmt3a and Dnmt3b was not observed. Deletion analysis showed that Dnmt1 recruitment was mediated by the PCNA-binding domain. These data point to a direct role of Dnmt1 in the restoration of epigenetic information during DNA repair.
Figures
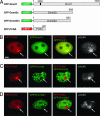
Fig. 1.
GFP-Dnmt1 and RFP-PCNA but not GFP-Dnmt3a and GFP-Dnmt3b1 colocalize with γ-H2AX at microirradiated sites. Wide-field fluorescence images of cotransfected C2C12 cells formaldehyde fixed 25–30 min after UVA laser microirradiation. Double strand breaks were detected with rabbit polyclonal antibodies against γ-H2AX. Arrows mark sites of irradiation. (A) Schematic representation of the fusion proteins. (B) GFP-Dnmt1 and RFP-PCNA accumulate at sites of DNA damage and colocalize with γ-H2AX. (C and D) GFP-Dnmt3a and GFP-Dnmt3b1 are bleached at irradiated sites and do not redistribute to sites of DNA damage after microirradiation. (Scale bar, 5 μm.)
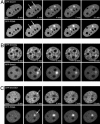
Fig. 2.
Dynamics of DNA methyltransferase recruitment to repair sites in living cells. Live cell imaging of microirradiated C2C12 cells in S phase coexpressing fluorescent fusion constructs of DNA methyltransferases and PCNA is shown. The constructs used were the same as depicted in Fig. 1 A except for the RFP-Dnmt1 construct where the GFP was replaced by monomeric RFP. Maximum projections of confocal midsections are shown and times after microirradiation are indicated. (A) A C2C12 cell in early to mid S phase coexpressing GFP-Dnmt1 and RFP-PCNA shows accumulation of RFP-PCNA and GFP-Dnmt1 at sites of DNA damage (arrows) as early as 1 min after irradiation. (B) An early S phase cell coexpressing GFP-Dnmt3a and RFP-Dnmt1 shows accumulation of RFP-Dnmt1 at the irradiated site (arrow), whereas GFP-Dnmt3a is bleached (arrow) and does not recover during the entire observation period. (C) An early S phase cell coexpressing GFP-Dnmt3b1 and RFP-PCNA shows bleaching of GFP-Dnmt3b1 (arrow) at the irradiation spot, whereas RFP-PCNA accumulates at this site (arrow). (Scale bars, 5 μm.)
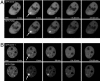
Fig. 3.
Recruitment of GFP-Dnmt1 and RFP-PCNA to repair sites in S and non-S phase C2C12 cells. The structure of the fusion proteins is depicted in Fig. 1 A. Live cell imaging of microirradiated C2C12 cells in G1 (A) and late S phase (B) coexpressing GFP-Dnmt1 and RFP-PCNA shows accumulation of RFP-PCNA and GFP-Dnmt1 at sites of DNA damage (arrows). Maximum projections of confocal midsections are shown and times after microirradiation are indicated. (Scale bars, 5 μm.)
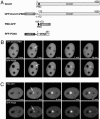
Fig. 4.
Recruitment of human Ligase 3 fused to GFP (GFP-Ligase 3) and RFP-PCNA or RFP-Dnmt1 to DNA repair sites after microirradiation. (A) Schematic representation of the fusion proteins. (B) Correct expression of the GFP-Ligase 3 construct was determined by Western blot analysis. (C and D) Live cell imaging of C2C12 cells coexpressing GFP-Ligase 3 and either RFP-PCNA (C) or RFP-Dnmt1 (D). After microirradiation with a 405-nm diode laser, GFP-Ligase 3, RFP-PCNA, and RFP-Dnmt1 accumulate at sites of DNA damage (arrows). One confocal midsection is shown and times after microirradiation are indicated. (Scale bars, 5 μm.)
Fig. 5.
Recruitment of Dnmt1 to DNA damage sites is mediated by PCNA. (A) Schematic representation of the fusion proteins. (B) Live cell imaging of a microirradiated C2C12 cell coexpressing GFP-Dnmt1ΔPBD and RFP-PCNA. Deletion of the PBD in GFP-Dnmt1ΔPBD abolishes recruitment to sites of DNA damage, whereas RFP-PCNA accumulates at these sites as seen before (arrow). (C) A late S phase cell coexpressing PBD-GFP and RFP-PCNA shows accumulation of both RFP-PCNA and PBD-GFP at sites of microirradiation (arrows). Maximum projections of confocal midsections are shown and times after microirradiation are indicated. (Scale bars, 5 μm.)
Similar articles
- Spatiotemporal dynamics of p21CDKN1A protein recruitment to DNA-damage sites and interaction with proliferating cell nuclear antigen.
Perucca P, Cazzalini O, Mortusewicz O, Necchi D, Savio M, Nardo T, Stivala LA, Leonhardt H, Cardoso MC, Prosperi E. Perucca P, et al. J Cell Sci. 2006 Apr 15;119(Pt 8):1517-27. doi: 10.1242/jcs.02868. Epub 2006 Mar 21. J Cell Sci. 2006. PMID: 16551699 - Recruitment of DNA repair synthesis machinery to sites of DNA damage/repair in living human cells.
Hashiguchi K, Matsumoto Y, Yasui A. Hashiguchi K, et al. Nucleic Acids Res. 2007;35(9):2913-23. doi: 10.1093/nar/gkm115. Epub 2007 Apr 16. Nucleic Acids Res. 2007. PMID: 17439963 Free PMC article. - Differential recruitment of DNA Ligase I and III to DNA repair sites.
Mortusewicz O, Rothbauer U, Cardoso MC, Leonhardt H. Mortusewicz O, et al. Nucleic Acids Res. 2006 Jul 19;34(12):3523-32. doi: 10.1093/nar/gkl492. Print 2006. Nucleic Acids Res. 2006. PMID: 16855289 Free PMC article. - Structural Basis of DNMT1 and DNMT3A-Mediated DNA Methylation.
Ren W, Gao L, Song J. Ren W, et al. Genes (Basel). 2018 Dec 11;9(12):620. doi: 10.3390/genes9120620. Genes (Basel). 2018. PMID: 30544982 Free PMC article. Review. - Function of heterochromatin protein 1 during DNA repair.
Bártová E, Malyšková B, Komůrková D, Legartová S, Suchánková J, Krejčí J, Kozubek S. Bártová E, et al. Protoplasma. 2017 May;254(3):1233-1240. doi: 10.1007/s00709-017-1090-3. Epub 2017 Feb 24. Protoplasma. 2017. PMID: 28236007 Review.
Cited by
- Epigenetic regulation of miR-17~92 contributes to the pathogenesis of pulmonary fibrosis.
Dakhlallah D, Batte K, Wang Y, Cantemir-Stone CZ, Yan P, Nuovo G, Mikhail A, Hitchcock CL, Wright VP, Nana-Sinkam SP, Piper MG, Marsh CB. Dakhlallah D, et al. Am J Respir Crit Care Med. 2013 Feb 15;187(4):397-405. doi: 10.1164/rccm.201205-0888OC. Epub 2013 Jan 10. Am J Respir Crit Care Med. 2013. PMID: 23306545 Free PMC article. - Cell division drives DNA methylation loss in late-replicating domains in primary human cells.
Endicott JL, Nolte PA, Shen H, Laird PW. Endicott JL, et al. Nat Commun. 2022 Nov 8;13(1):6659. doi: 10.1038/s41467-022-34268-8. Nat Commun. 2022. PMID: 36347867 Free PMC article. - Inhibition of DNA Methylation Impairs Synaptic Plasticity during an Early Time Window in Rats.
Muñoz P, Estay C, Díaz P, Elgueta C, Ardiles ÁO, Lizana PA. Muñoz P, et al. Neural Plast. 2016;2016:4783836. doi: 10.1155/2016/4783836. Epub 2016 Jul 14. Neural Plast. 2016. PMID: 27493805 Free PMC article. - MBD4 and MLH1 are required for apoptotic induction in xDNMT1-depleted embryos.
Ruzov A, Shorning B, Mortusewicz O, Dunican DS, Leonhardt H, Meehan RR. Ruzov A, et al. Development. 2009 Jul;136(13):2277-86. doi: 10.1242/dev.032227. Development. 2009. PMID: 19502488 Free PMC article. - Integrated analysis of DNA methylation and gene expression reveals specific signaling pathways associated with platinum resistance in ovarian cancer.
Li M, Balch C, Montgomery JS, Jeong M, Chung JH, Yan P, Huang TH, Kim S, Nephew KP. Li M, et al. BMC Med Genomics. 2009 Jun 8;2:34. doi: 10.1186/1755-8794-2-34. BMC Med Genomics. 2009. PMID: 19505326 Free PMC article.
References
- Friedberg, E. C. (2003) Nature 421, 436–440. - PubMed
- Hoeijmakers, J. H. (2001) Nature 411, 366–374. - PubMed
- Jaenisch, R. & Bird, A. (2003) Nat. Genet. 33, Suppl., 245–254. - PubMed
- Robertson, K. D. (2002) Oncogene 21, 5361–5379. - PubMed
- Leonhardt, H. & Cardoso, M. C. (2000) J. Cell Biochem., Suppl. 35, 78–83. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous