Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation - PubMed (original) (raw)
Natural antisense transcripts with coding capacity in Arabidopsis may have a regulatory role that is not linked to double-stranded RNA degradation
Chih-Hung Jen et al. Genome Biol. 2005.
Abstract
Background: Overlapping transcripts in antisense orientation have the potential to form double-stranded RNA (dsRNA), a substrate for a number of different RNA-modification pathways. One prominent route for dsRNA is its breakdown by Dicer enzyme complexes into small RNAs, a pathway that is widely exploited by RNA interference technology to inactivate defined genes in transgenic lines. The significance of this pathway for endogenous gene regulation remains unclear.
Results: We have examined transcription data for overlapping gene pairs in Arabidopsis thaliana. On the basis of an analysis of transcripts with coding regions, we find the majority of overlapping gene pairs to be convergently overlapping pairs (COPs), with the potential for dsRNA formation. In all tissues, COP transcripts are present at a higher frequency compared to the overall gene pool. The probability that both the sense and antisense copy of a COP are co-transcribed matches the theoretical value for coexpression under the assumption that the expression of one partner does not affect the expression of the other. Among COPs, we observe an over-representation of spliced (intron-containing) genes (90%) and of genes with alternatively spliced transcripts. For loci where antisense transcripts overlap with sense transcript introns, we also find a significant bias in favor of alternative splicing and variation of polyadenylation.
Conclusion: The results argue against a predominant RNA degradation effect induced by dsRNA formation. Instead, our data support alternative roles for dsRNAs. They suggest that at least for a subgroup of COPs, antisense expression may induce alternative splicing or polyadenylation.
Figures
Figure 1
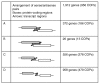
A comparison of the arrangements of overlapping gene pairs in Arabidopsis thaliana. A and A' label the start and end of the sense transcript, B' and B label the start and end of the antisense transcript. The total number of genes involved in group 1, 2 and 3 is 2,157, of which 2,147 are unique; the remaining 10 comprise four genes that are members of both group 1 and group 2 pairs, five genes that are members of both group 1 and group 3 pairs, and 1 gene that is a member of both a group 1 and group 3 pair.
Figure 2
The organization of convergent overlapping gene pairs with respect to the protein coding capacity of the sense and antisense transcripts.
Figure 3
Illustration of the distance between the end of the antisense transcript and the last intron-exon boundary of the sense transcript. Negative values refer to a termination of the antisense transcript 5' to the intron-exon boundary.
Similar articles
- Sense and antisense transcripts of convergent gene pairs in Arabidopsis thaliana can share a common polyadenylation region.
Zubko E, Kunova A, Meyer P. Zubko E, et al. PLoS One. 2011 Feb 2;6(2):e16769. doi: 10.1371/journal.pone.0016769. PLoS One. 2011. PMID: 21311762 Free PMC article. - Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis.
Jin H, Vacic V, Girke T, Lonardi S, Zhu JK. Jin H, et al. BMC Mol Biol. 2008 Jan 14;9:6. doi: 10.1186/1471-2199-9-6. BMC Mol Biol. 2008. PMID: 18194570 Free PMC article. - Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana.
Wang XJ, Gaasterland T, Chua NH. Wang XJ, et al. Genome Biol. 2005;6(4):R30. doi: 10.1186/gb-2005-6-4-r30. Epub 2005 Mar 15. Genome Biol. 2005. PMID: 15833117 Free PMC article. - Dual RNAs in plants.
Bardou F, Merchan F, Ariel F, Crespi M. Bardou F, et al. Biochimie. 2011 Nov;93(11):1950-4. doi: 10.1016/j.biochi.2011.07.028. Epub 2011 Jul 31. Biochimie. 2011. PMID: 21824505 Review. - Alternative polyadenylation of antisense RNAs and flowering time control.
Hornyik C, Duc C, Rataj K, Terzi LC, Simpson GG. Hornyik C, et al. Biochem Soc Trans. 2010 Aug;38(4):1077-81. doi: 10.1042/BST0381077. Biochem Soc Trans. 2010. PMID: 20659007 Review.
Cited by
- Magnaporthe grisea infection triggers RNA variation and antisense transcript expression in rice.
Gowda M, Venu RC, Li H, Jantasuriyarat C, Chen S, Bellizzi M, Pampanwar V, Kim H, Dean RA, Stahlberg E, Wing R, Soderlund C, Wang GL. Gowda M, et al. Plant Physiol. 2007 May;144(1):524-33. doi: 10.1104/pp.107.095653. Epub 2007 Mar 9. Plant Physiol. 2007. PMID: 17351054 Free PMC article. - Abscisic acid response element binding factor 1 is required for establishment of Arabidopsis seedlings during winter.
Sharma PD, Singh N, Ahuja PS, Reddy TV. Sharma PD, et al. Mol Biol Rep. 2011 Nov;38(8):5147-59. doi: 10.1007/s11033-010-0664-3. Epub 2010 Dec 24. Mol Biol Rep. 2011. PMID: 21181499 - Classification of and detection techniques for RNAi-induced effects in GM plants.
Diaz C, Ayobahan SU, Simon S, Zühl L, Schiermeyer A, Eilebrecht E, Eilebrecht S. Diaz C, et al. Front Plant Sci. 2025 Mar 7;16:1535384. doi: 10.3389/fpls.2025.1535384. eCollection 2025. Front Plant Sci. 2025. PMID: 40123947 Free PMC article. Review. - Prediction of trans-antisense transcripts in Arabidopsis thaliana.
Wang H, Chua NH, Wang XJ. Wang H, et al. Genome Biol. 2006;7(10):R92. doi: 10.1186/gb-2006-7-10-r92. Epub 2006 Oct 13. Genome Biol. 2006. PMID: 17040561 Free PMC article. - The Functions of RNA-Dependent RNA Polymerases in Arabidopsis.
Willmann MR, Endres MW, Cook RT, Gregory BD. Willmann MR, et al. Arabidopsis Book. 2011;9:e0146. doi: 10.1199/tab.0146. Epub 2011 Jul 31. Arabidopsis Book. 2011. PMID: 22303271 Free PMC article.
References
- Boi S, Solda G, Tenchini ML. Shedding light on the dark side of the genome: overlapping genes in higher eukaryotes. Curr Genomics. 2004;5:509–524. doi: 10.2174/1389202043349020. - DOI
- Hastings ML, Milcarek C, Martincic K, Peterson ML, Munroe SH. Expression of the thyroid hormone receptor gene, erbAα, in B lymphocytes: alternative mRNA processing is independent of differentiation but correlates with antisense RNA levels. Nucleic Acids Res. 1997;25:4296–4300. doi: 10.1093/nar/25.21.4296. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases