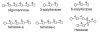Automated interpretation of MS/MS spectra of oligosaccharides - PubMed (original) (raw)
Automated interpretation of MS/MS spectra of oligosaccharides
Haixu Tang et al. Bioinformatics. 2005 Jun.
Abstract
Motivation: The emerging glycomics and glycoproteomics projects aim to characterize all forms of glycoproteins in different tissues and organisms. Tandem mass spectrometry (MS/MS) is the key experimental methodology for high-throughput glycan identification and characterization. Fragmentation of glycans from high energy collision-induced dissociation generates ions from glycosidic as well as internal cleavages. The cross-ring ions resulting from internal cleavages provide additional information that is important to reveal the type of linkage between monosaccharides. This information, however, is not incorporated into the current programs for analyzing glycan mass spectra. As a result, they can rarely distinguish from the mass spectra isomeric oligosaccharides, which have the same saccharide composition but different types of sequences, branches or linkages.
Results: In this paper, we describe a novel algorithm for glycan characterization using MS/MS. This algorithm consists of three steps. First, we develop a scoring scheme to identify potential bond linkages between monosaccharides, based on the appearance pattern of cross-ring ions. Next, we use a dynamic programming algorithm to determine the most probable oligosaccharide structures from the mass spectrum. Finally, we re-evaluate these oligosaccharide structures, taking into account the double fragmentation ions. We also show the preliminary results of testing our algorithm on several MS/MS spectra of oligosaccharides.
Availability: The program GLYCH is available upon request from the authors.
Figures
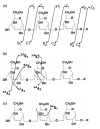
Fig. 1.
The structure of oligosaccharides. (a) The open (acyclic) form structure of glucose, an epimer of hexose, one of the monosaccharides, i.e. the basic unit of an oligosaccharide. (b) The cyclic form structure of glucose. The carbon atoms in the ring are not shown. (c) A diglucose, consisting of two glucoses forming a 1-4 glycosidic bond. (d) A tetraglucose, consisting of four glucoses with a branching. (e) The symbolic representation of the tetraglucose shown in (d). The numbers show the linkage types.
Fig. 2.
The fragmentation patterns of oligosaccharides. (a) The Y/b and Z/C ions are from the glycosidic cleavages. (b) The cross-ring cleavages create X/A ions. (c) Since the middle unit of the triglucose is linked to two other glucoses, some cleavages create fragment ions with small masses that are not close to the corresponding Y/b ions, e.g. 0,4_X_1. These ions will be ignored in our program.
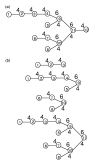
Fig. 3.
The PRMs of an oligosacchrides. (a) An oligosaccharide consisting of 12 glucoses. The glucose residues are indexed according to the partial order of nodes of the tree, from leafs to the root. For any two residues ri and rj, if ri is within the subtree rooted by rj, i must be indexed smaller than j.(b) The subtrees rooted by residues _r_3, _r_9, _r_10, _r_11.
Fig. 4.
The structures of the oligosaccharides used for testing in this paper.
Similar articles
- A Machine Learning Based Approach to de novo Sequencing of Glycans from Tandem Mass Spectrometry Spectrum.
Kumozaki S, Sato K, Sakakibara Y. Kumozaki S, et al. IEEE/ACM Trans Comput Biol Bioinform. 2015 Nov-Dec;12(6):1267-74. doi: 10.1109/TCBB.2015.2430317. IEEE/ACM Trans Comput Biol Bioinform. 2015. PMID: 26671799 - Cross-Ring Fragmentation Patterns in the Tandem Mass Spectra of Underivatized Sialylated Oligosaccharides and Their Special Suitability for Spectrum Library Searching.
De Leoz MLA, Simón-Manso Y, Woods RJ, Stein SE. De Leoz MLA, et al. J Am Soc Mass Spectrom. 2019 Mar;30(3):426-438. doi: 10.1007/s13361-018-2106-8. Epub 2018 Dec 18. J Am Soc Mass Spectrom. 2019. PMID: 30565163 Free PMC article. - Methods in enzymology: O-glycosylation of proteins.
Peter-Katalinić J. Peter-Katalinić J. Methods Enzymol. 2005;405:139-71. doi: 10.1016/S0076-6879(05)05007-X. Methods Enzymol. 2005. PMID: 16413314 Review. - Bioinformatics protocols in glycomics and glycoproteomics.
Tang H, Mayampurath A, Yu CY, Mechref Y. Tang H, et al. Curr Protoc Protein Sci. 2014 Apr 1;76:2.15.1-2.15.7. doi: 10.1002/0471140864.ps0215s76. Curr Protoc Protein Sci. 2014. PMID: 24692013 Review.
Cited by
- Insights into Bioinformatic Applications for Glycosylation: Instigating an Awakening towards Applying Glycoinformatic Resources for Cancer Diagnosis and Therapy.
Muthu M, Chun S, Gopal J, Anthonydhason V, Haga SW, Jacintha Prameela Devadoss A, Oh JW. Muthu M, et al. Int J Mol Sci. 2020 Dec 8;21(24):9336. doi: 10.3390/ijms21249336. Int J Mol Sci. 2020. PMID: 33302373 Free PMC article. Review. - Identification and accurate quantitation of biological oligosaccharide mixtures.
Strum JS, Kim J, Wu S, De Leoz ML, Peacock K, Grimm R, German JB, Mills DA, Lebrilla CB. Strum JS, et al. Anal Chem. 2012 Sep 18;84(18):7793-801. doi: 10.1021/ac301128s. Epub 2012 Aug 29. Anal Chem. 2012. PMID: 22897719 Free PMC article. - Sweetening the pot: adding glycosylation to the biomarker discovery equation.
Drake PM, Cho W, Li B, Prakobphol A, Johansen E, Anderson NL, Regnier FE, Gibson BW, Fisher SJ. Drake PM, et al. Clin Chem. 2010 Feb;56(2):223-36. doi: 10.1373/clinchem.2009.136333. Epub 2009 Dec 3. Clin Chem. 2010. PMID: 19959616 Free PMC article. Review. - Software for automated interpretation of mass spectrometry data from glycans and glycopeptides.
Woodin CL, Maxon M, Desaire H. Woodin CL, et al. Analyst. 2013 May 21;138(10):2793-803. doi: 10.1039/c2an36042j. Analyst. 2013. PMID: 23293784 Free PMC article. Review. - Automated Glycan Sequencing from Tandem Mass Spectra of N-Linked Glycopeptides.
Yu CY, Mayampurath A, Zhu R, Zacharias L, Song E, Wang L, Mechref Y, Tang H. Yu CY, et al. Anal Chem. 2016 Jun 7;88(11):5725-32. doi: 10.1021/acs.analchem.5b04858. Epub 2016 May 23. Anal Chem. 2016. PMID: 27111718 Free PMC article.
References
- Bafna V, Edwards N. On de novo interpretation of peptide mass spectra. Proceedings of Research in Computational Biology (RECOMB).2003. pp. 9–18.
- Bandeira N, Tang H, Bafna V, Pevzner PA. Shotgun protein sequencing by tandem mass spectra assembly. Anal. Chem. 2004;76:7221–7233. - PubMed
- Chen T, Kao MY, Tepel M, Rush J, Church GM. A dynamic programming approach to de novo peptide sequencing via tandem mass spectrometry. J. Comput. Biol. 2001;8:325–337. - PubMed
- Cooper CA, Gasteiger E, Packer NH. GlycoMod—a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics. 2001;1:340–349. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials