The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex - PubMed (original) (raw)
The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex
Qun He et al. Genes Dev. 2005.
Abstract
The COP9 signalosome (CSN) promotes the function of SCF-type cullin-based ubiquitin ligase complexes in vivo. Paradoxically, removal of the Nedd8 modification of cullins by CSN inhibits the ubiquitin ligase activity of SCF complexes in vitro. Ubiquitination-mediated degradation of the Neurospora circadian clock protein FREQUENCY (FRQ) is critical for clock function. Ubiquitination of FRQ requires FWD-1, the substrate-recruiting subunit of an SCF complex. Here we show that disruption of a subunit of CSN (csn-2) impairs the degradation of FRQ and compromises its normal circadian expression. A FRQ-independent oscillator drives conidiation in the csn-2 mutant, resulting in a 2-d conidiation rhythm that persists in constant darkness (DD), constant light (LL), light-to-dark (LD) transitions, and temperature cycles. Strikingly, the levels of FWD-1 are drastically reduced in csn-2 mutant, explaining the impaired degradation of FRQ. Reduction of FWD-1 levels in the mutant requires its F-box, suggesting that its degradation is due to autoubiquitination. In addition, SKP-1 and CUL-1 of the SCF(FWD-1) complex are also unstable in the mutant. Therefore, our results establish an important role of CSN in the circadian clock of Neurospora. Our findings also reconcile the CSN paradox and suggest that a major function of CSN is to maintain the stability of SCF ubiquitin ligases in vivo.
Figures
Figure 1.
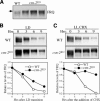
Disruption of csn-2 leads to growth and developmental phenotypes and hyperneddylation of Cul-1. (A) The wild-type and csn-2 mutant strains grown in minimal slants. Note that the csn-2 strains produced significantly less aerial hyphae and conidia than the wild-type strain. (B) Southern blot analysis showing the disruption of the endogenous csn-2 gene in the csn-2 mutants. The genomic DNA was digested by EcoRI, and a DNA fragment containing the CSN-2 ORF was used as the probe. (C) The expression profile of Myc-CUL-1 in the wild-type and csn-2KO strains. Western blot analysis was performed using the c-Myc antibody.
Figure 2.
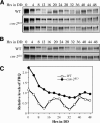
Degradation of FRQ is impaired in the csn-2 mutant. (A) Western blot analysis showing the expression of FRQ in the wild-type and csn-2KO strains. Cultures were grown in LL. Note that FRQ levels are high and hyperphosphorylated. The arrow indicates the hyperphosphorylated FRQ species. (B,C) Degradation of FRQ after a LD transition (B) or the addition of CHX (10 μg/mg). (C) The bottom panels show the densitometric results of the Western blot data. Four independent experiments were performed and similar data were obtained.
Figure 3.
Loss of normal circadian expression of FRQ under normal growth (A) and nutrient depletion conditions (B,C) in the csn-2 mutant. Cultures were harvested at the indicated hours in DD for 2 d. For experiments described in A, the cultures were grown in liquid media for 48–60 h before harvest, whereas in B they were grown 72–84 h to achieve nutrient depletion. The densitometric results of the Western blot data in B are shown in C. For experiments described in A, four independent experiments were performed and the results of two representative experiments are shown. For B, the experiments were repeated twice and similar results were obtained in both experiments.
Figure 4.
Abnormal conidiation rhythms of the csn-2 mutant in DD (A), LD (B), LL (C), and temperature cycles (D), or at different constant temperatures in DD (E). At least six replicates were examined under each condition and representative race tube results are shown. The black lines mark the growth fronts every 24 h.
Figure 5.
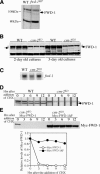
The FWD-1 levels are drastically reduced in the csn-2 mutant, probably due to its rapid autoubiquitination-mediated degradation. (A) Western blot analysis showing the expression of FWD-1 in the wild-type and fwd-1RIP strains. The arrow indicates the FWD-1 specific band in the wild-type strain. The asterisk indicates an unspecific cross-reacted protein band recognized by our FWD-1 antiserum. (B) The expression of FWD-1 in the wild-type and csn-2KO strains after 2 or 3 d of growth in liquid medium. (C) Northern blot analysis showing the expression of fwd-1 mRNA in the wild-type and csn-2KO strains. (D) Western blot analysis showing the degradation of FWD-1 after the addition of CHX. Similar results were obtained in three independent experiments. (E) Degradation of Myc-FWD-1 or Myc-FWD-1ΔF (lacking its F-box) expressed in the csn-2 mutants after the addition of CHX. The densitometric analysis of the results is shown in the bottom panel.
Figure 6.
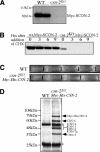
The components of the SCFFWD-1 complex, SKP1 and CUL-1, are also unstable in the csn-2 mutant. (A) Race tube assays showing the complementation of the circadian conidiation rhythm defect of the fwd-1RIP strain by the expression Myc-His-FWD-1. (B) Silver-stained SDS-PAGE gel showing biochemical purification of Myc-His-FWD-1 and its associated proteins. A wild-type strain was used as the negative control. Myc-His-FWD-1 and SKP-1 were identified by mass spectrometry analysis. (C) Immunoprecipitation (IP) assays showing the association between FWD-1 and Myc-CUL-1. The extracts of the wt,Myc-Cul-1 strain were immunoprecipitated by the preimmune serum (PI) or the FWD-1 antiserum, followed by Western blot analysis using the c-Myc antibody. (D,E) Degradation of Myc-CUL-1 (D) or Myc-SKP-1 (E) in the wild-type strain or the csn-2 mutant. Similar results were obtained in two independent experiments. The densitometric analyses of the results in D and E are shown in F and G, respectively.
Figure 7.
SCON-2 is rapidly degraded in the csn-2 mutant, resulting in its drastically reduced expression level. (A) Expression level of Myc-SCON-2 in the wild-type and csn-2KO strains in LL. Two independent transformants of each strain were used. (B) Western blot analysis showing the degradation of Myc-SCON-2 in the wild-type and csn-2KO strains. (C) Race tube assays showing the complementation of the circadian conidiation rhythm defect of the cns-2KO strain by the expression Myc-His-CSN-2. (D) Silver-stained SDS-PAGE gel showing biochemical purification of the Neurospora CSN. A wild-type strain was used as the negative control. CSN subunits identified by mass spectrometry analysis are indicated. Asterisks indicate the two IgG bands.
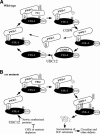
Figure 8.
A model of the SCFFWD-1 cycle in a wild-type (A) strain and in csn mutants (B).
Similar articles
- Degradation of the Neurospora circadian clock protein FREQUENCY through the ubiquitin-proteasome pathway.
He Q, Liu Y. He Q, et al. Biochem Soc Trans. 2005 Nov;33(Pt 5):953-6. doi: 10.1042/BST20050953. Biochem Soc Trans. 2005. PMID: 16246019 - Neurospora COP9 signalosome integrity plays major roles for hyphal growth, conidial development, and circadian function.
Zhou Z, Wang Y, Cai G, He Q. Zhou Z, et al. PLoS Genet. 2012;8(5):e1002712. doi: 10.1371/journal.pgen.1002712. Epub 2012 May 10. PLoS Genet. 2012. PMID: 22589747 Free PMC article. - Role of individual subunits of the Neurospora crassa CSN complex in regulation of deneddylation and stability of cullin proteins.
Wang J, Hu Q, Chen H, Zhou Z, Li W, Wang Y, Li S, He Q. Wang J, et al. PLoS Genet. 2010 Dec 2;6(12):e1001232. doi: 10.1371/journal.pgen.1001232. PLoS Genet. 2010. PMID: 21151958 Free PMC article. - Mammalian COP9 signalosome.
Kato JY, Yoneda-Kato N. Kato JY, et al. Genes Cells. 2009 Nov;14(11):1209-25. doi: 10.1111/j.1365-2443.2009.01349.x. Epub 2009 Oct 22. Genes Cells. 2009. PMID: 19849719 Review. - COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases.
Cope GA, Deshaies RJ. Cope GA, et al. Cell. 2003 Sep 19;114(6):663-71. doi: 10.1016/s0092-8674(03)00722-0. Cell. 2003. PMID: 14505567 Review.
Cited by
- The COP9 signalosome and cullin-RING ligases in the heart.
Wang X, Martin DS. Wang X, et al. Am J Cardiovasc Dis. 2015 Mar 20;5(1):1-18. eCollection 2015. Am J Cardiovasc Dis. 2015. PMID: 26064789 Free PMC article. Review. - The Arabidopsis F-box protein CORONATINE INSENSITIVE1 is stabilized by SCFCOI1 and degraded via the 26S proteasome pathway.
Yan J, Li H, Li S, Yao R, Deng H, Xie Q, Xie D. Yan J, et al. Plant Cell. 2013 Feb;25(2):486-98. doi: 10.1105/tpc.112.105486. Epub 2013 Feb 5. Plant Cell. 2013. PMID: 23386265 Free PMC article. - Transcription factor CCG-8 plays a pivotal role in azole adaptive responses of Neurospora crassa by regulating intracellular azole accumulation.
Xue W, Yin Y, Ismail F, Hu C, Zhou M, Cao X, Li S, Sun X. Xue W, et al. Curr Genet. 2019 Jun;65(3):735-745. doi: 10.1007/s00294-018-0924-7. Epub 2019 Jan 2. Curr Genet. 2019. PMID: 30603874 - Neddylation and CAND1 independently stimulate SCF ubiquitin ligase activity in Candida albicans.
Sela N, Atir-Lande A, Kornitzer D. Sela N, et al. Eukaryot Cell. 2012 Jan;11(1):42-52. doi: 10.1128/EC.05250-11. Epub 2011 Nov 11. Eukaryot Cell. 2012. PMID: 22080453 Free PMC article. - A plant cytorhabdovirus modulates locomotor activity of insect vectors to enhance virus transmission.
Gao DM, Qiao JH, Gao Q, Zhang J, Zang Y, Xie L, Zhang Y, Wang Y, Fu J, Zhang H, Han C, Wang XB. Gao DM, et al. Nat Commun. 2023 Sep 16;14(1):5754. doi: 10.1038/s41467-023-41503-3. Nat Commun. 2023. PMID: 37717061 Free PMC article.
References
- Akten B., Jauch, E., Genova, G.K., Kim, E.Y., Edery, I., Raabe, T., and Jackson, F.R. 2003. A role for CK2 in the Drosophila circadian oscillator. Nat. Neurosci. 6: 251–257. - PubMed
- Aronson B., Johnson, K., Loros, J.J., and Dunlap, J.C. 1994a. Negative feedback defining a circadian clock: Autoregulation in the clock gene frequency. Science 263: 1578–1584. - PubMed
- Busch S., Eckert, S.E., Krappmann, S., and Braus, G.H. 2003. The COP9 signalosome is an essential regulator of development in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 49: 717–730. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous