Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma - PubMed (original) (raw)
Comparative Study
Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma
Raju R Raval et al. Mol Cell Biol. 2005 Jul.
Abstract
Defective function of the von Hippel-Lindau (VHL) tumor suppressor ablates proteolytic regulation of hypoxia-inducible factor alpha subunits (HIF-1alpha and HIF-2alpha), leading to constitutive activation of hypoxia pathways in renal cell carcinoma (RCC). Here we report a comparative analysis of the functions of HIF-1alpha and HIF-2alpha in RCC and non-RCC cells. We demonstrate common patterns of HIF-alpha isoform transcriptional selectivity in VHL-defective RCC that show consistent and striking differences from patterns in other cell types. We also show that HIF-alpha isoforms display unexpected suppressive interactions in RCC cells, with enhanced expression of HIF-2alpha suppressing HIF-1alpha and vice-versa. In VHL-defective RCC cells, we demonstrate that the protumorigenic genes encoding cyclin D1, transforming growth factor alpha, and vascular endothelial growth factor respond specifically to HIF-2alpha and that the proapoptotic gene encoding BNip3 responds positively to HIF-1alpha and negatively to HIF-2alpha, indicating that HIF-1alpha and HIF-2alpha have contrasting properties in the biology of RCC. In keeping with this, HIF-alpha isoform-specific transcriptional selectivity was matched by differential effects on the growth of RCC as tumor xenografts, with HIF-1alpha retarding and HIF-2alpha enhancing tumor growth. These findings indicate that therapeutic approaches to targeting of the HIF system, at least in this setting, will need to take account of HIF isoform-specific functions.
Figures
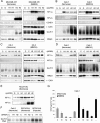
FIG. 1.
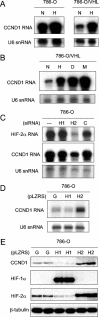
Regulation of cyclin D1 (CCND1) in 786-O cells. (A and B) RNase protection assay of CCND1 mRNA and U6 snRNA (loading control) in 786-O (PRC3) and 786-O/VHL (WT-8) after exposure to normoxia (N), hypoxia (H), 100 μM desferrioxamine (D), or 1 mM MMOG (M) for 18 h. (C) RNase protection assay of HIF-2α mRNA and CCND1 mRNA in 786-O/VHL (WT-8) after treatment for 48 h with Oligofectamine alone (−), HIF-1α-directed siRNA (H1), HIF-2α-directed siRNA (H2), or control siRNA (C). (D) RNase protection assay of CCND1 mRNA and U6 snRNA in 786-O cells infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-1α (H1), or HIF-2α (H2). (E) Immunoblots, with the indicated antibodies, of whole-cell lysates from 786-O cells that were infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-1α (H1), or HIF-2α (H2). Two independent experiments for each condition are shown in panel E.
FIG. 2.
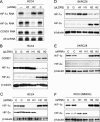
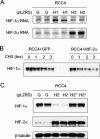
Interaction between HIF-1α and HIF-2α and effects on cyclin D1 expression. (A) RNase protection assay of HIF-1α, HIF-2α, and CCND1 mRNAs (U6 snRNA, loading control) in RCC4 cells after treatment for 48 h with Oligofectamine alone (−), HIF-1α-directed siRNA (H1), HIF-2α-directed siRNA (H2), or control siRNA (C). (B and D) Immunoblots, with the indicated antibodies, of whole-cell lysates from RCC4 and SKRC28 cells that were infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-1α (H1), or HIF-2α (H2). (C and E) Immunoblots for HIF-1α, HIF-2α, and β-tubulin of RCC4 and SKRC28 whole-cell lysates after treatment for 48 h with a control siRNA (C), HIF-1α siRNA (H1), or HIF-2α siRNA (H2). (F) Immunoblots for HIF-1α, HIF-2α, and β-tubulin of RCC4 whole-cell lysates treated with the previously indicated siRNAs and then exposed to 1 mM MMOG for 18 h. Two independent experiments for each condition are shown in panels B, C, D, E, and F.
FIG. 3.
Analysis of suppression of HIF-1α by HIF-2α in RCC4 cells. (A) RNase protection assay of HIF-1α and HIF-2α mRNAs in RCC4 cells infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-1α (H1), or HIF-2α (H2). (B) Representative immunoblot for HIF-1α of RCC4 whole-cell lysates which were infected with retroviral supernatants made from pLZRS containing GFP alone or HIF-2α and then treated with 100 μM cycloheximide (CHX) for 0, 1, 2, or 3 h. Note that to aid densitometric analysis of signal decay, more extract was loaded from cells infected with retroviruses expressing HIF-2α than GFP alone so as to obtain sufficient signal intensity at t = 0. (C) Immunoblots for HIF-1α, HIF-2α, and β-tubulin of whole-cell lysates from RCC4 cells infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-2α (H2), or a HIF-2α DNA binding domain mutant (H2*). Two independent experiments for each condition are shown in panels A and C.
FIG. 4.
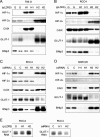
HIF-α isoform transcriptional specificity in 786-O, RCC4, and SKRC28 cells. (A and B) Immunoblots for HIF-1α, HIF-2α, CAIX, GLUT-1, and BNip3 of 786-O and RCC4 whole-cell lysates that were infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-1α (H1), or HIF-2α (H2). (C and D) Immunoblots, with the indicated antibodies, of whole-cell lysates from RCC4 and SKCR28 cells after treatment for 48 h with a control siRNA (C), HIF-1α siRNA (H1), or HIF-2α siRNA (H2). (E) RNase protection assay of GLUT-1 mRNA in RCC4 cells infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-1α (H1), or HIF-2α (H2) and after treatment for 48 h with a control siRNA (C), HIF-1α siRNA (H1), or HIF-2α siRNA (H2). Two independent experiments for each condition are shown in panels A, B, C, and D.
FIG. 5.
Unusual patterns of HIF-α-dependent gene expression in RCC cells. (A) VEGF secretion by RCC4 and SKRC28 cells as determined by ELISA of medium supernatant. Cells were treated with siRNAs directed against a control sequence (C), HIF-1α (H1), HIF-2α (H2), both HIF-1α and HIF-2α (B), or Oligofectamine alone (−). VEGF levels were normalized to cell number. Experiments were performed in triplicate at least three times, and error bars correspond to 1 standard deviation. (B) Expression levels of TGF-α in 786-O, RCC4, and SKRC28 cell lysates as determined by ELISA. Cells were treated with siRNAs directed against a control sequence (C), HIF-1α (H1), HIF-2α (H2), both HIF-1α and HIF-2α (B), or Oligofectamine alone (−). TGF-α expression levels in the cell lysates were normalized to total cellular protein content. Experiments were performed in triplicate at least three times, and error bars correspond to 1 standard deviation. (C) Immunoblots for HIF-1α, HIF-2α, and β-tubulin of whole-cell lysates from 786-O/VHL and A498/VHL cells after exposure to normoxia (N) or 1 mM MMOG (M) for 18 h. Two independent experiments for each condition are shown in panel C.
FIG. 6.
HIF-α isoform transcriptional selectivity in MDA-MB 435, Caki-1, and HK-2 cells. (A and B) Immunoblots, with the indicated antibodies, of MDA-MB 435 and Caki-1 whole-cell lysates that were infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-1α (H1), or HIF-2α (H2) and then exposed to normoxia or 1 mM MMOG for the final 18 h. (C and D) Immunoblots, with the indicated antibodies, of whole-cell lysates from HK-2 and Caki-1 cells after treatment for 48 h with control siRNA (C), HIF-1α siRNA (H1), or HIF-2α siRNA (H2) and then exposed to normoxia or 1 mM MMOG for the final 18 h. (E) Immunoblots for HIF-1α and HIF-2α of RCC4/VHL whole-cell lysates that were infected with retroviral supernatants made from pLZRS containing GFP alone (G), HIF-1α (H1), or HIF-2α (H2). Two independent experiments for each condition are shown in panels A, B, C, D, and E. (F) RNase protection assay of GLUT-1 mRNA in Caki-1, RCC4/VHL (Normoxia), and RCC4/VHL (Hypoxia) cells after treatment for 48 h with a control siRNA (C), HIF-1α siRNA (H1), or HIF-2α siRNA (H2). (G) Secreted levels of VEGF in Caki-1 as determined by ELISA of medium supernatant. Cells were treated with siRNAs directed against a control sequence (C), HIF-1α (H1), HIF-2α (H2), both HIF-1α and HIF-2α (B), or Oligofectamine alone (−) and then exposed to normoxia or 1 mM MMOG for the final 18 h. VEGF levels were normalized to cell number. Experiments were performed in triplicate at least three times, and error bars correspond to 1 standard deviation.
FIG. 7.
Immunohistochemical analysis for HIF-α and transcriptional targets in sections from the kidney of a patient with VHL disease. Immunohistochemical analysis, with the indicated antibodies, of serial sections of representative lesions. Panels: A, an early multicellular lesion; B, a renal cyst; C, an overt clear-cell RCC. Final magnifications: A, ×360; B and C, ×240.
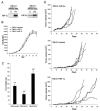
FIG. 8.
Effect of overexpression of HIF-1α or HIF-2α on growth of 786-O cells as monolayers or tumor xenografts. (A) Immunoblots for HIF-1α and HIF-2α of whole-cell lysates from 786-O polyclonal pools of cells that were selected with G418 after infection with retroviral supernatants made from pBMN-Neo containing an empty cassette (LacZ spliced out) as a control (C), HIF-1α (H1), or HIF-2α (H2). (B) Growth of selected polyclonal pools of 786-O cells stably infected with the indicated retroviruses. Growth was measured under standard tissue culture conditions for 9 days, and there was no significant difference in the proliferation rate between the groups. Error bars indicate standard deviations. (C) Tumor weights approximately 5.5 weeks after subcutaneous injection of polyclonal pools of 786-O cells stably infected with the indicated retroviruses. There were 14 tumors analyzed for each group, and error bars indicate 1 standard error. Two-tailed, unpaired Student t tests comparing each HIF-α overexpressing group to the control group were performed, and statistically significant difference is indicated by asterisks for P < 0.005. (D) Growth curves indicated as tumor volume over a 90-day period after subcutaneous injection of polyclonal pools of 786-O cells stably infected with the indicated retroviruses. Five mice with single tumors were analyzed for each group.
Similar articles
- Contrasting effects on HIF-1alpha regulation by disease-causing pVHL mutations correlate with patterns of tumourigenesis in von Hippel-Lindau disease.
Clifford SC, Cockman ME, Smallwood AC, Mole DR, Woodward ER, Maxwell PH, Ratcliffe PJ, Maher ER. Clifford SC, et al. Hum Mol Genet. 2001 May 1;10(10):1029-38. doi: 10.1093/hmg/10.10.1029. Hum Mol Genet. 2001. PMID: 11331613 - Hypoxia, Hypoxia-inducible Transcription Factors, and Renal Cancer.
Schödel J, Grampp S, Maher ER, Moch H, Ratcliffe PJ, Russo P, Mole DR. Schödel J, et al. Eur Urol. 2016 Apr;69(4):646-657. doi: 10.1016/j.eururo.2015.08.007. Epub 2015 Aug 19. Eur Urol. 2016. PMID: 26298207 Free PMC article. Review. - The HIF pathway: implications for patterns of gene expression in cancer.
Wykoff CC, Pugh CW, Harris AL, Maxwell PH, Ratcliffe PJ. Wykoff CC, et al. Novartis Found Symp. 2001;240:212-25; discussion 225-31. doi: 10.1002/0470868716.ch15. Novartis Found Symp. 2001. PMID: 11727931 Review.
Cited by
- USP7 depletion potentiates HIF2α degradation and inhibits clear cell renal cell carcinoma progression.
Tu R, Ma J, Chen Y, Kang Y, Ren D, Cai Z, Zhang R, Pan Y, Liu Y, Da Y, Xu Y, Yu Y, Wang D, Wang J, Dong Y, Lu X, Zhang C. Tu R, et al. Cell Death Dis. 2024 Oct 15;15(10):749. doi: 10.1038/s41419-024-07136-0. Cell Death Dis. 2024. PMID: 39406703 Free PMC article. - Toward a CRISPR-based mouse model of _Vhl_-deficient clear cell kidney cancer: Initial experience and lessons learned.
Stransky LA, Gao W, Schmidt LS, Bi K, Ricketts CJ, Ramesh V, James A, Difilippantonio S, Ileva L, Kalen JD, Karim B, Jeon A, Morgan T, Warner AC, Turan S, Unite J, Tran B, Choudhari S, Zhao Y, Linn DE, Yun C, Dhandapani S, Parab V, Pinheiro EM, Morris N, He L, Vigeant SM, Pignon JC, Sticco-Ivins M, Signoretti S, Van Allen EM, Linehan WM, Kaelin WG Jr. Stransky LA, et al. Proc Natl Acad Sci U S A. 2024 Oct 8;121(41):e2408549121. doi: 10.1073/pnas.2408549121. Epub 2024 Oct 4. Proc Natl Acad Sci U S A. 2024. PMID: 39365820 Free PMC article. - Molecular chaperones: Guardians of tumor suppressor stability and function.
Heritz JA, Backe SJ, Mollapour M. Heritz JA, et al. Oncotarget. 2024 Oct 1;15:679-696. doi: 10.18632/oncotarget.28653. Oncotarget. 2024. PMID: 39352796 Free PMC article. Review. - Next-Cell Hypothesis: Mechanism of Obesity-Associated Carcinogenesis.
Engin AB, Engin A. Engin AB, et al. Adv Exp Med Biol. 2024;1460:727-766. doi: 10.1007/978-3-031-63657-8_25. Adv Exp Med Biol. 2024. PMID: 39287871 Review. - Saturation genome editing maps the functional spectrum of pathogenic VHL alleles.
Buckley M, Terwagne C, Ganner A, Cubitt L, Brewer R, Kim DK, Kajba CM, Forrester N, Dace P, De Jonghe J, Shepherd STC, Sawyer C, McEwen M, Diederichs S, Neumann-Haefelin E, Turajlic S, Ivakine EA, Findlay GM. Buckley M, et al. Nat Genet. 2024 Jul;56(7):1446-1455. doi: 10.1038/s41588-024-01800-z. Epub 2024 Jul 5. Nat Genet. 2024. PMID: 38969834 Free PMC article.
References
- Baba, M., S. Hirai, H. Yamada-Okabe, K. Hamada, H. Tabuchi, K. Kobayashi, K. Kondo, M. Yoshida, A. Yamashita, T. Kishida, N. Nakaigawa, Y. Nagashima, Y. Kubota, M. Yao, and S. Ohno. 2003. Loss of von Hippel-Lindau protein causes cell density dependent deregulation of cyclin D1 expression through hypoxia-inducible factor. Oncogene 22:2728-2738. - PubMed
- Bindra, R. S., J. R. Vasselli, R. Stearman, W. M. Linehan, and R. D. Klausner. 2002. VHL-mediated hypoxia regulation of cyclin D1 in renal carcinoma cells. Cancer Res. 62:3014-3019. - PubMed
- Bui, M. H., D. Seligson, K. R. Han, A. J. Pantuck, F. J. Dorey, Y. Huang, S. Horvath, B. C. Leibovich, S. Chopra, S. Y. Liao, E. Stanbridge, M. I. Lerman, A. Palotie, R. A. Figlin, and A. S. Belldegrun. 2003. Carbonic anhydrase IX is an independent predictor of survival in advanced renal clear cell carcinoma: implications for prognosis and therapy. Clin. Cancer Res. 9:802-811. - PubMed
- Carmeliet, P., Y. Dor, J. M. Herbert, D. Fukumura, K. Brusselmans, M. Dewerchin, M. Neeman, F. Bono, R. Abramovitch, P. Maxwell, C. J. Koch, P. Ratcliffe, L. Moons, R. K. Jain, D. Collen, E. Keshert, and E. Keshet. 1998. Role of HIF-1α in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 394:485-490. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials