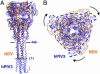Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein - PubMed (original) (raw)
Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein
Hsien-Sheng Yin et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Class I viral fusion proteins share common mechanistic and structural features but little sequence similarity. Structural insights into the protein conformational changes associated with membrane fusion are based largely on studies of the influenza virus hemagglutinin in pre- and postfusion conformations. Here, we present the crystal structure of the secreted, uncleaved ectodomain of the paramyxovirus, human parainfluenza virus 3 fusion (F) protein, a member of the class I viral fusion protein group. The secreted human parainfluenza virus 3 F forms a trimer with distinct head, neck, and stalk regions. Unexpectedly, the structure reveals a six-helix bundle associated with the postfusion form of F, suggesting that the anchor-minus ectodomain adopts a conformation largely similar to the postfusion state. The transmembrane anchor domains of F may therefore profoundly influence the folding energetics that establish and maintain a metastable, prefusion state.
Figures
Fig. 1.
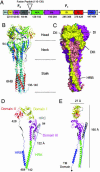
Structure of the hPIV3 solF0 protein. (A) Schematic of the domain structure of the hPIV3 solF0 protein. Domain regions are indicated with hPIV3 sequence numbers shown below and with colors corresponding to those used in Fig. 1_D_.(B) Ribbon diagram of the hPIV3 solF0 trimer. The three chains are colored similarly from blue (N terminus) to red (C terminus). Residues 95-135 are disordered in all chains. Residue 94 is labeled in one chain and residues 136-140 at the base of the stalk are ordered in one chain because of crystal packing interactions. (C) Surface representation of the solF0 trimer. Each chain is a different color and domains I-III and HRB for one chain (yellow) are indicated by the DI, DII, DIII, and HRB labels. One radial channel is readily apparent below domains I and II of the yellow chain and above domain III of the red chain. (D) Ribbon diagram of the solF0 protein monomer colored by domain. The direct distance within one monomer between residue 94 at the end of HRC and residue 142 at the base of the stalk region is 122 Å. (E) Ribbon diagram of the monomer rotated by 90°, indicating the width and height of the solF0 monomer. An arrow at the C terminus of the HRB segment points toward the likely position of the TM anchor domain that would be present in the full-length protein.
Fig. 2.
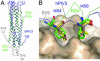
Structural comparison with SV5 and hRSV F1 core structures. (A) Overlay of the hPIV3 solF0, SV5 F1 core, and hRSV core structures. The SV5 N1/C1 peptide complex is shown in pale red. The hRSV N57/C45 complex is shown in pale green. (B) Comparison of hydrophobic pocket interactions of hPIV3 residues I454 and I456 with those in SV5 and hRSV. The hPIV3 residues are shown in yellow, with the backbone trace of the C-terminal segment shown in blue. The hRSV residues are shown in pale green, and the SV5 residues are shown in pale red.
Fig. 3.
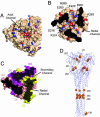
Structural features of the hPIV3 head region and location of fusion mutations. (A) Top view of the hPIV3 trimer showing the axial channel. The surface is shown with positions of potentially charged atoms shown in either red (for the carboxylate oxygens of Asp and Glu residues) or blue (for the nitrogen atoms of Lys, Arg, and His residues). (B) Slice through the head region of the hPIV3 solF0 trimer at the position of the radial channel. The surface is colored as in A, and the positions of charged residues are indicated. (C) Surface representation of a narrow, secondary radial channel located between monomers at the DII:DI packing interface. Chains are colored yellow, pale red, and magenta. (D) Locations of residues that affect paramyxovirus F protein fusion activity. Residues are shown as yellow cpk representations, and only one of the three is labeled for clarity. See Table 2 for additional information on the mutants and their effects.
Fig. 4.
Structural comparison with the proteolytic fragment of NDV F. (A) Overlay of trimers by using the coiled coil for the superposition. hPIV3 solF0 is shown in blue, and NDV F is shown in orange. The NDV F stalk terminates above the six-helix bundle observed in hPIV3 solF0, terminating at the indicated hPIV3 residues (171 and 448). (B) Top view of the superimposed trimers, colored as in A. The Head domain of hPIV3 solF0 is rotated clockwise relative to the head region of NDV F. The orientation differences are due to significant changes in the packing of the individual F protein domains I-III.
Similar articles
- Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein.
Welch BD, Liu Y, Kors CA, Leser GP, Jardetzky TS, Lamb RA. Welch BD, et al. Proc Natl Acad Sci U S A. 2012 Oct 9;109(41):16672-7. doi: 10.1073/pnas.1213802109. Epub 2012 Sep 10. Proc Natl Acad Sci U S A. 2012. PMID: 23012473 Free PMC article. - Inhibiting Human Parainfluenza Virus Infection by Preactivating the Cell Entry Mechanism.
Bottom-Tanzer SF, Rybkina K, Bell JN, Alabi CA, Mathieu C, Lu M, Biswas S, Vasquez M, Porotto M, Melero JA, Más V, Moscona A. Bottom-Tanzer SF, et al. mBio. 2019 Feb 19;10(1):e02900-18. doi: 10.1128/mBio.02900-18. mBio. 2019. PMID: 30782664 Free PMC article. - Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation.
Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Yin HS, et al. Nature. 2006 Jan 5;439(7072):38-44. doi: 10.1038/nature04322. Nature. 2006. PMID: 16397490 Free PMC article. - Structural basis of viral invasion: lessons from paramyxovirus F.
Lamb RA, Jardetzky TS. Lamb RA, et al. Curr Opin Struct Biol. 2007 Aug;17(4):427-36. doi: 10.1016/j.sbi.2007.08.016. Epub 2007 Sep 17. Curr Opin Struct Biol. 2007. PMID: 17870467 Free PMC article. Review. - Structural and functional specificity of Influenza virus haemagglutinin and paramyxovirus fusion protein anchoring peptides.
Kordyukova L. Kordyukova L. Virus Res. 2017 Jan 2;227:183-199. doi: 10.1016/j.virusres.2016.09.014. Epub 2016 Oct 20. Virus Res. 2017. PMID: 27773768 Review.
Cited by
- Structures of the Foamy virus fusion protein reveal an unexpected link with the F protein of paramyxo- and pneumoviruses.
Fernández I, Bontems F, Brun D, Coquin Y, Goverde CA, Correia BE, Gessain A, Buseyne F, Rey FA, Backovic M. Fernández I, et al. Sci Adv. 2024 Oct 11;10(41):eado7035. doi: 10.1126/sciadv.ado7035. Epub 2024 Oct 11. Sci Adv. 2024. PMID: 39392890 Free PMC article. - Human parainfluenza virus 3 field strains undergo extracellular fusion protein cleavage to activate entry.
Stearns K, Lampe G, Hanan R, Marcink T, Niewiesk S, Sternberg SH, Greninger AL, Porotto M, Moscona A. Stearns K, et al. mBio. 2024 Nov 13;15(11):e0232724. doi: 10.1128/mbio.02327-24. Epub 2024 Oct 9. mBio. 2024. PMID: 39382296 Free PMC article. - Systematic computer-aided disulfide design as a general strategy to stabilize prefusion class I fusion proteins.
Gonzalez KJ, Yim KC, Blanco JCG, Boukhvalova MS, Strauch EM. Gonzalez KJ, et al. Front Immunol. 2024 Jul 24;15:1406929. doi: 10.3389/fimmu.2024.1406929. eCollection 2024. Front Immunol. 2024. PMID: 39114655 Free PMC article. - Structural basis for potent neutralization of human respirovirus type 3 by protective single-domain camelid antibodies.
Johnson NV, van Scherpenzeel RC, Bakkers MJG, Ramamohan AR, van Overveld D, Le L, Langedijk JPM, Kolkman JA, McLellan JS. Johnson NV, et al. Nat Commun. 2024 Jun 27;15(1):5458. doi: 10.1038/s41467-024-49757-1. Nat Commun. 2024. PMID: 38937429 Free PMC article. - Functional and structural basis of human parainfluenza virus type 3 neutralization with human monoclonal antibodies.
Suryadevara N, Otrelo-Cardoso AR, Kose N, Hu YX, Binshtein E, Wolters RM, Greninger AL, Handal LS, Carnahan RH, Moscona A, Jardetzky TS, Crowe JE Jr. Suryadevara N, et al. Nat Microbiol. 2024 Aug;9(8):2128-2143. doi: 10.1038/s41564-024-01722-w. Epub 2024 Jun 10. Nat Microbiol. 2024. PMID: 38858594
References
- Dutch, R. E., Jardetzky, T. S. & Lamb, R. A. (2000) Biosci. Rep. 20, 597-612. - PubMed
- Jardetzky, T. S. & Lamb, R. A. (2004) Nature 427, 307-308. - PubMed
- Lamb, R. A. & Kolakofsky, D. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott Williams & Wilkins, Philadelphia), 4th Ed., pp. 1305-1340.
- Skehel, J. J. & Wiley, D. C. (2000) Annu. Rev. Biochem. 69, 531-569. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources