p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation - PubMed (original) (raw)
p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation
Vikram Devgan et al. Genes Dev. 2005.
Abstract
In keratinocytes, the cyclin/CDK inhibitor p21(WAF1/Cip1) is a direct transcriptional target of Notch1 activation; loss of either the p21 or Notch1 genes expands stem cell populations and facilitates tumor development. The Notch1 tumor-suppressor function was associated with down-regulation of Wnt signaling. Here, we show that suppression of Wnt signaling by Notch1 activation is mediated, at least in part, by down-modulation of Wnts gene expression. p21 is a negative regulator of Wnts transcription downstream of Notch1 activation, independently of effects on the cell cycle. More specifically, expression of the Wnt4 gene is under negative control of endogenous p21 both in vitro and in vivo. p21 associates with the E2F-1 transcription factor at the Wnt4 promoter and causes curtailed recruitment of c-Myc and p300, and histone hypoacetylation at this promoter. Thus, p21 acts as a selective negative regulator of transcription and links the Notch and Wnt signaling pathways in keratinocyte growth control.
Figures
Figure 1.
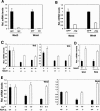
Notch1 activation suppresses Wnts gene expression. (A) Down-modulation of Wnts gene expression by activated Notch1. Primary mouse keratinocytes were infected with a recombinant adenovirus expressing the constitutive activated form of Notch1 (NIC) or an adenovirus expressing GFP (GFP) for 24 h. mRNA levels for Wnt3 and Wnt4 were quantified by real-time RT-PCR. Values are expressed as relative arbitrary units after normalization for GAPDH mRNA levels. (B) Down-modulation of Wnts gene expression in response to increased Jagged1 expression. Primary keratinocytes were infected with a recombinant adenovirus expressing full-length Jagged 1 (Jag) or the Ad-GFP control (GFP) for 24 h, followed by mRNA quantification of Wnt3 and Wnt4 as in A. (C) Differential down-modulation of Wnts gene expression upon induction of differentiation of Notch1+/+ versus _Notch1_-/- keratinocytes. Primary keratinocytes derived from mice with the Notch1 gene flanked by loxP sites were infected with a Cre-expressing adenovirus to delete the endogenous Notch1 gene as previously described (Rangarajan et al. 2001). Parallel cultures of the same cells were infected with Ad-GFP as controls. Three days after infection, keratinocytes were induced to differentiate by exposure to elevated extracellular calcium for 3 d (3d) or 6 d (6d). Total RNAs from Notch1+/+ versus _Notch1_-/- cells were analyzed by real-time RT-PCR for Wnt3 and Wnt4 expression as in A and B. (D) Increased Wnts expression in the epidermis of mice with an induced deletion of the Notch1 gene. Total RNA was prepared from the isolated epidermis of mice with the Notch1 gene flanked by loxP sites and carrying a keratinocyte-specific Cre transgene versus control Cre-negative littermates (Rangarajan et al. 2001). Wnt3 and Wnt4 mRNA levels were quantified by real-time RT-PCR as before. (E) Differential down-modulation of Wnts gene expression upon induction of differentiation of RBP-Jk+/+ versus _RBP-Jk_-/- keratinocytes. Primary keratinocytes derived from mice with the RBP-Jk gene flanked by loxP sites were infected with a Cre-expressing adenovirus to delete the endogenous RBP-Jk gene as previously described (Mammucari et al. 2005). Parallel cultures of the same cells were infected with Ad-GFP as controls. Three days after infection, keratinocytes were induced to differentiate by exposure to elevated extracellular calcium for 3 d (3d) or 6 d (6d). Total RNAs from RBP-Jk+/+ versus _RBP-Jk_-/- cells were analyzed by real-time RT-PCR for Wnt3 and Wnt4 expression as in A-D.
Figure 2.
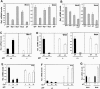
Notch1 suppresses Wnt signaling by down-modulating Wnts gene expression. (A-C) Differential regulation of p21, integrin α6, and K1 expression by Notch1 in control versus K14-Wnt3 transgenic keratinocytes. Primary keratinocytes derived from K14-Wnt3 transgenic mice (Millar et al. 1999) versus transgenic-negative littermate controls were infected with the Ad-NIC (NIC) versus Ad-GFP (GFP) viruses, followed by mRNA quantification of p21, integrin α6 and K1 gene, respectively, by real-time RT-PCR. Values are expressed as relative folds of expression, after normalization for GAPDH mRNA levels. (D) Differential down-regulation of activated-β-catenin levels by activated Notch1 in control versus K14-Wnt3 transgenic keratinocytes. Primary keratinocytes derived from K14-Wnt3 transgenic mice versus transgenic-negative littermate controls were infected with the Ad-NIC (NIC) versus Ad-GFP (GFP) viruses. Total cell extracts were analyzed by immunoblotting with antibodies specific for the unphosphorylated activated form of β-catenin (van Noort et al. 2002), total-β-catenin or β-actin. (E) Differential down-regulation of activated-β-catenin levels by activated Notch1 in control versus _Wnt4_-overexpressing keratinocytes. Primary keratinocytes were infected with a Wnt4 transducing (retro-Wnt4) versus vector control (retro-pLNCX) retroviruses. Two days after infection, keratinocytes were subjected to selection for G418 resistance, followed by infection with the Ad-NIC (NIC) versus Ad-GFP (GFP) viruses. Total cell extracts were analyzed by immunoblotting for levels of activated β-catenin as in D.
Figure 3.
p21, like Hes-1, is a negative regulator of Wnts gene expression. (A) Hes-1 and p21WAF1/Cip1 are negative regulators of Wnts gene expression. Primary keratinocytes were infected with adenoviruses expressing the Hes-1, Hey-1, Hey-2, or p21 proteins for 24 h at a multiplicity of 100, followed by mRNA quantification of Wnt3 and Wnt4 as before. (B) Additive suppressive effects of Hes-1 and p21 on Wnts gene expression. Primary keratinocytes were infected with the adenoviruses expressing Hes-1 (Ad-Hes-1) or p21 (Ad-p21F) viruses either alone or in combination at a multiplicity of 50. Ad-GFP (GFP) was used as a control and added to the Ad-Hes-1 or Ad-p21F viruses when they were used alone, to ensure that cells received, in all cases, the same amount of viral particles (total multiplicity of infection: 100). Quantification of Wnt3 and Wnt4 mRNA expression by real-time RT-PCR was carried out as before. (C) Similar induction of Hes-1 expression by activated Notch1 in p21+/+ versus _p21_-/- keratinocytes. Primary keratinocytes derived from p21+/+ and _p21_-/- mice of the same genetic background (Sencar) were infected with the Ad-NIC (NIC) versus Ad-GFP (GFP) viruses. mRNA levels of Hes-1 were determined by real-time RT-PCR as before. Values are expressed as arbitrary units relative to Ad-GFP-infected control cells. (D) Differential suppression of Wnts gene expression by activated Notch1 in p21+/+ versus _p21_-/- keratinocytes. The same samples utilized in C were analyzed for levels of Wnt3 and Wnt4 mRNA expression by real-time RT-PCR. Values are expressed as arbitrary units relative, in each case, to Ad-GFP-infected control cells. (E,F) Differential expression of Wnt3 and Wnt4 gene expression upon induction of differentiation of p21+/+ versus _p21_-/- keratinocytes. Primary keratinocytes derived from p21+/+ and _p21_-/- mice were either kept under growing conditions or induced to differentiate by exposure to elevated extracellular calcium for 3 d (3d) or 6 d (6d). Wnts mRNA levels were quantified by real-time RT-PCR as before. Values are indicated in arbitrary units relative to untreated p21+/+ cells. (G) Increased Wnt4 mRNA expression in the epidermis of _p21_-/- mice. Total RNA was prepared from the separated epidermis of p21+/+ versus _p21_-/- mice and Wnt3 and Wnt4 mRNA levels were quantified as before.
Figure 4.
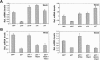
Increased p21 expression suppresses Wnts gene expression independently of effects on the cell cycle and at the transcription level. (A) Down-modulation of Wnts expression by p21 un-linked from the cell cycle. Primary keratinocytes were infected with adenoviruses expressing GFP (GFP), full-length p21 protein (p21F), the p21 N domain (p21N), p16INK4a (p16), and p27Kip1 (p27) for 24 h, followed by Wnt3 and Wnt4 mRNA quantification as before. (B) Counteracting effects of histone deacetylase inhibition on Wnts suppression by p21. Primary keratinocytes were infected with the Ad-GFP control or the p21-expressing adenovirus (p21F) with or without concomitant treatment with the histone deacetylase inhibitor M334 (5 μM, an amide analog of Trichostatin A [Jung et al. 1999]) plus/minus an antagonist of this compound, ITSA1 (10 μM, inhibitor of Trichostatin A 1 (Koeller et al. 2003). Wnt3 and Wnt4 mRNA levels were quantified by real-time RT-PCR as before.
Figure 5.
p21 binds to the endogenous Wnt4 promoter in association with E2F-1. (A) Map of the Wnt4 promoter region with indication of the E2F-1-binding sites (open squares) and the position of the oligonucleotide primers utilized for the ChIP analysis described below (nucleotides -6839 to -6718 [1]; nucleotides +21 to +121 [2] relative to TATA box). (B) Specific binding of endogenous E2F-1 to the TATA box-proximal region of the Wnt4 gene. Primary keratinocytes were infected with the Ad-GFP or Ad-p21F adenoviruses for 24 h. Cells were analyzed by ChIP with antibodies specific for E2F-1 or nonimmune controls, followed by real-time PCR amplification of the indicated regions of the Wnt4 gene. Chromatin preparations prior to the immunoprecipitation reaction were similarly processed and analyzed as controls for input DNA. (C) Specific binding of p21 to the TATA box-proximal region of the Wnt4 gene. Primary keratinocytes were infected with the Ad-GFP or Ad-p21F adenoviruses and analyzed by ChIP with antibodies against the p21 protein and corresponding controls as in the previous experiments. Immunoprecipitates were analyzed by real-time PCR for the indicated regions of the Wnt4 gene as well as for the E2F-1-binding region of the PCNA gene (nucleotides -398 to -270 relative to the AUG). (D) Association of the p21 and E2F-1 proteins at the TATA box-proximal region of the Wnt4 gene. Primary keratinocytes were infected with the Ad-GFP or Ad-p21F adenoviruses and subjected to the ChIP procedure with sequential immunoprecipitation with antibodies against the E2F-1 and p21 proteins or corresponding nonimmune controls. Immunoprecipitates were analyzed by real-time PCR for the indicated regions of the Wnt4 gene.
Figure 6.
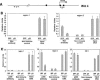
Increased p21 expression curtails recruitment of c-Myc and p300 to the Wnt4 promoter. (A) Map of the Wnt4 gene, with indication of the c-Myc-binding sites (solid squares) and the position of the oligonucleotide primers utilized for the ChIP analysis (the same as in Fig. 5A). (B) Decreased recruitment of the c-Myc protein to the TATA box-proximal region of the Wnt4 gene by increased p21 expression. Primary keratinocytes were infected with the Ad-GFP or p21F adenoviruses for 24 h and analyzed by ChIP with antibodies against the c-Myc protein and corresponding controls as in the previous experiments. Immunoprecipitates were analyzed by real-time PCR for the indicated regions of the Wnt4 gene. (C) Decreased recruitment of the p300 protein to the TATA box-proximal region of the Wnt4 gene by increased p21 expression. Primary keratinocytes were infected with the Ad-GFP or Ad-p21F adenoviruses for 24 h and analyzed by ChIP with antibodies against the p300 protein and corresponding controls as in the previous experiments. Immunoprecipitates were analyzed by real-time PCR for the indicated regions of the Wnt4 gene, as well as for the TATA box-proximal region of the IGF-1 gene (nucleotides -242 to -141 relative to the AUG).
Figure 7.
Increased p21 expression causes histone hypoacetylation at specific regions of the Wnt4 promoter. (A) Map of the Wnt4 gene encompassing the TATA box, with indication of the position of the oligonucleotide primers utilized for the ChIP analysis performed below (nucleotides -6839 to -6718 [1]; nucleotides -1175 to -1075 [1a]; nucleotides +21 to +121 [2]; nucleotides +1329 to +1430 [3] relative to TATA box). (B) Specific down-modulation of histone 4 acetylation levels at the TATA box-proximal region of the Wnt4 gene by exogenously increased p21 expression. Primary keratinocytes were infected with the Ad-GFP or Ad-p21F adenoviruses for 24 h. Cells were analyzed by ChIP with antibodies specific for the acetylated form of histone 4 (at amino acids 4, 7, 11, 15), followed by real-time PCR amplification of the indicated regions of the Wnt4 gene. (C) Differential control of histone 4 acetylation levels at the TATA box-proximal region of the Wnt4 gene by activated Notch1 as a function of endogenous p21. Primary keratinocytes from p21+/+ and _p21_-/- mice were infected with the Ad-GFP or Ad-NIC viruses for 24 h and analyzed by ChIP with antibodies specific for the acetylated form of histone 4 and corresponding nonimmune controls as in the previous figures.
Similar articles
- Integration of Notch 1 and calcineurin/NFAT signaling pathways in keratinocyte growth and differentiation control.
Mammucari C, Tommasi di Vignano A, Sharov AA, Neilson J, Havrda MC, Roop DR, Botchkarev VA, Crabtree GR, Dotto GP. Mammucari C, et al. Dev Cell. 2005 May;8(5):665-76. doi: 10.1016/j.devcel.2005.02.016. Dev Cell. 2005. PMID: 15866158 - Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation.
Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, Aster JC, Krishna S, Metzger D, Chambon P, Miele L, Aguet M, Radtke F, Dotto GP. Rangarajan A, et al. EMBO J. 2001 Jul 2;20(13):3427-36. doi: 10.1093/emboj/20.13.3427. EMBO J. 2001. PMID: 11432830 Free PMC article. - Notch signaling induces cell cycle arrest in small cell lung cancer cells.
Sriuranpong V, Borges MW, Ravi RK, Arnold DR, Nelkin BD, Baylin SB, Ball DW. Sriuranpong V, et al. Cancer Res. 2001 Apr 1;61(7):3200-5. Cancer Res. 2001. PMID: 11306509 - A dynamic model of keratinocyte stem cell renewal and differentiation: role of the p21WAF1/Cip1 and Notch1 signaling pathways.
Okuyama R, LeFort K, Dotto GP. Okuyama R, et al. J Investig Dermatol Symp Proc. 2004 Sep;9(3):248-52. doi: 10.1111/j.1087-0024.2004.09308.x. J Investig Dermatol Symp Proc. 2004. PMID: 15369220 Review. - Effects of histone acetylation and DNA methylation on p21( WAF1) regulation.
Fang JY, Lu YY. Fang JY, et al. World J Gastroenterol. 2002 Jun;8(3):400-5. doi: 10.3748/wjg.v8.i3.400. World J Gastroenterol. 2002. PMID: 12046058 Free PMC article. Review.
Cited by
- Stromal epigenetic dysregulation is sufficient to initiate mouse prostate cancer via paracrine Wnt signaling.
Zong Y, Huang J, Sankarasharma D, Morikawa T, Fukayama M, Epstein JI, Chada KK, Witte ON. Zong Y, et al. Proc Natl Acad Sci U S A. 2012 Dec 11;109(50):E3395-404. doi: 10.1073/pnas.1217982109. Epub 2012 Nov 26. Proc Natl Acad Sci U S A. 2012. PMID: 23184966 Free PMC article. - Does Notch play a tumor suppressor role across diverse squamous cell carcinomas?
Zhang M, Biswas S, Qin X, Gong W, Deng W, Yu H. Zhang M, et al. Cancer Med. 2016 Aug;5(8):2048-60. doi: 10.1002/cam4.731. Epub 2016 May 26. Cancer Med. 2016. PMID: 27228302 Free PMC article. Review. - Sulforaphane induction of p21(Cip1) cyclin-dependent kinase inhibitor expression requires p53 and Sp1 transcription factors and is p53-dependent.
Chew YC, Adhikary G, Wilson GM, Xu W, Eckert RL. Chew YC, et al. J Biol Chem. 2012 May 11;287(20):16168-78. doi: 10.1074/jbc.M111.305292. Epub 2012 Mar 15. J Biol Chem. 2012. PMID: 22427654 Free PMC article. - The Role of the Cyclin Dependent Kinase Inhibitor p21cip1/waf1 in Targeting Cancer: Molecular Mechanisms and Novel Therapeutics.
Al Bitar S, Gali-Muhtasib H. Al Bitar S, et al. Cancers (Basel). 2019 Sep 30;11(10):1475. doi: 10.3390/cancers11101475. Cancers (Basel). 2019. PMID: 31575057 Free PMC article. Review. - CDKN1C negatively regulates RNA polymerase II C-terminal domain phosphorylation in an E2F1-dependent manner.
Ma Y, Chen L, Wright GM, Pillai SR, Chellappan SP, Cress WD. Ma Y, et al. J Biol Chem. 2010 Mar 26;285(13):9813-9822. doi: 10.1074/jbc.M109.091496. Epub 2010 Jan 27. J Biol Chem. 2010. PMID: 20106982 Free PMC article.
References
- Adnane J., Jackson, R.J., Nicosia, S.V., Cantor, A.B., Pledger, W.J., and Sebti, S.M. 2000. Loss of p21WAF1/CIP1 accelerates Ras oncogenesis in a transgenic/knockout mammary cancer model. Oncogene 19: 5338-5347. - PubMed
- Artavanis-Tsakonas S., Rand, M.D., and Lake, R.J. 1999. Notch signaling: Cell fate control and signal integration in development. Science 284: 770-776. - PubMed
- Axelrod J.D., Matsuno, K., Artavanis-Tsakonas, S., and Perrimon, N. 1996. Interaction between Wingless and Notch signaling pathways mediated by dishevelled. Science 271: 1826-1832. - PubMed
- Cheng T., Rodrigues, N., Shen, H., Yang, Y., Dombkowski, D., Sykes, M., and Scadden, D.T. 2000. Hematopoietic stem cell quiescence maintained by p21(cip1/waf1). Science 287: 1804-1808. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AR39190/AR/NIAMS NIH HHS/United States
- CA16038/CA/NCI NIH HHS/United States
- R01 CA073796/CA/NCI NIH HHS/United States
- R01 AR039190/AR/NIAMS NIH HHS/United States
- P01 CA016038/CA/NCI NIH HHS/United States
- CA73796/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous