Distribution and intensity of constraint in mammalian genomic sequence - PubMed (original) (raw)
Comparative Study
Distribution and intensity of constraint in mammalian genomic sequence
Gregory M Cooper et al. Genome Res. 2005 Jul.
Abstract
Comparisons of orthologous genomic DNA sequences can be used to characterize regions that have been subject to purifying selection and are enriched for functional elements. We here present the results of such an analysis on an alignment of sequences from 29 mammalian species. The alignment captures approximately 3.9 neutral substitutions per site and spans approximately 1.9 Mbp of the human genome. We identify constrained elements from 3 bp to over 1 kbp in length, covering approximately 5.5% of the human locus. Our estimate for the total amount of nonexonic constraint experienced by this locus is roughly twice that for exonic constraint. Constrained elements tend to cluster, and we identify large constrained regions that correspond well with known functional elements. While constraint density inversely correlates with mobile element density, we also show the presence of unambiguously constrained elements overlapping mammalian ancestral repeats. In addition, we describe a number of elements in this region that have undergone intense purifying selection throughout mammalian evolution, and we show that these important elements are more numerous than previously thought. These results were obtained with Genomic Evolutionary Rate Profiling (GERP), a statistically rigorous and biologically transparent framework for constrained element identification. GERP identifies regions at high resolution that exhibit nucleotide substitution deficits, and measures these deficits as "rejected substitutions". Rejected substitutions reflect the intensity of past purifying selection and are used to rank and characterize constrained elements. We anticipate that GERP and the types of analyses it facilitates will provide further insights and improved annotation for the human genome as mammalian genome sequence data become richer.
Figures
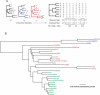
Figure 1.
Overview of GERP. (A) Each column of the compressed alignment (corresponding to each base of the human sequence) is analyzed independently. Number of substitution events is inferred, giving “observed” values (see Methods); the “expected” rate for each column is determined by summing the branches of the neutral tree that remain after removing species with a gap character (compare the black, red, and blue neutral trees with the correspondingly colored expected rates). Candidate constrained regions are identified as consecutive columns of observed rates smaller than the expected rates (black boxes). Nearby candidates are merged (gray box) across a limited number of unconstrained columns. Finally, each candidate is scored as the sum of the deviations from expectation at each column, collectively termed as “rejected substitutions.” (B) Neutral tree for the complete set of species analyzed here (see Methods); the tree is rooted arbitrarily for display purposes only, and analyses are performed using an unrooted tree. Primates are in green, non-primate placental mammals are in red, and marsupials are in blue.
Figure 2.
Confidence and sensitivity of GERP as a function of the rejected substitution threshold used to identify constrained elements. (A) Number of constrained element bases identified in the real alignment (solid line) and permuted alignments (dashed line). (B) Confidence is defined as the number of constrained element bases in the actual alignment divided by the sum of the constrained element bases in the actual and permuted alignments (see Methods). In A and B, the curves indicated with “+” and “–” characters result from analyses using a neutral rate estimate that is 10% greater or less, respectively, than the estimate of 3.85 neutral subs/site. A vertical black line marks the RS score threshold of 8.5 (corresponding to a confidence of ∼95%). (C) The fraction of exons that overlap at least one constrained element (solid line), and the fraction of exonic bases within a constrained element (dashed line). (D) Cumulative frequencies of the sizes of constrained elements at an RS of 8.5 or greater, with permuted alignment elements (heavy dashed line), exclusively nonexonic constrained elements (solid line), and exonic elements (light dashed line).
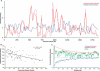
Figure 3.
Constrained elements tend to cluster, and this clustering is inversely correlated with repetitive element density. (A) Densities of constrained elements (red) and repetitive elements (blue) along the length of the human CFTR locus. Densities are determined for consecutive, nonoverlapping 25-kb windows, and each window is normalized by the locus-wide average. The solid red line corresponds to constrained elements identified with a merging tolerance of one unconstrained column, as opposed to six unconstrained columns for the dashed line (Fig. 1A; see Methods). (B) Regional constrained element density vs. repetitive element density. The values for each 25-kb window used in A are shown. The equation and trendline correspond to a simple linear regression model relating the two variables, with an R2 value of 0.32. (C) Constrained element density as a function of distance from various features (see Methods); (solid red line) constrained elements with a merging tolerance of one unconstrained column; (dashed red line) constrained elements with a merging tolerance of six unconstrained columns; (green line) exons; (blue line) repeats. Note that the behavior of the red lines very near the origin is a result of the fact that a pair of elements cannot be within the “merge distance” of each other (see Methods).
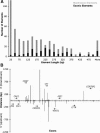
Figure 4.
Description of large constrained regions in the CFTR locus. (A) Sizes of constrained elements identified with a merging tolerance of six unconstrained columns, with the length in base pairs of each bin along the _x_-axis and the count for each bin along the _y_-axis. Bins are divided according to those elements that overlap exons (black) and those that do not (gray). (B) Large, non-coding constrained elements that overlap coding exons (see Methods). Each coding exon in the region is displayed in ascending order along the _x_-axis according to human genome coordinates. Exons are boxed according to which gene they belong, and transcription orientation of each gene is shown with an arrow. Note that in this format, the _left_-most exon is the first coding exon for all of those genes transcribed to the right, while the opposite is true for genes transcribed toward the left. The distance that the associated noncoding constrained elements extend away from the individual exons is plotted along the _y_-axis. Positive values are indicated for the 3′ direction, and negative values for the 5′ direction.
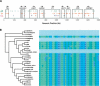
Figure 5.
Ultraconserved and mobile-element derived constrained elements. (A) The locations of three types of features are shown along the genomic coordinates of the human locus as follows: green squares indicate the locations of ancestral repeats that overlap constrained elements; orange triangles correspond to exons; and circles correspond to the ultraconserved elements (Table 2), broken down according to those that overlap exons (red), and those that do not (blue). RefSeq genes and their transcriptional orientation are marked by boxes and arrows, respectively. (B) A small alignment region corresponding to an ancestral repeat region (part of an L3 element) overlapping a constrained element scoring >100 rejected substitutions. Nucleotides are color-coded, and gaps are indicated in gray (the fully gapped placental species are missing data). The displayed region corresponds to positions 495,140–495,181 of the human sequence (with the first base of the locus being position 1).
Similar articles
- Population genetic models of GERP scores suggest pervasive turnover of constrained sites across mammalian evolution.
Huber CD, Kim BY, Lohmueller KE. Huber CD, et al. PLoS Genet. 2020 May 29;16(5):e1008827. doi: 10.1371/journal.pgen.1008827. eCollection 2020 May. PLoS Genet. 2020. PMID: 32469868 Free PMC article. - Evidence for turnover of functional noncoding DNA in mammalian genome evolution.
Smith NG, Brandström M, Ellegren H. Smith NG, et al. Genomics. 2004 Nov;84(5):806-13. doi: 10.1016/j.ygeno.2004.07.012. Genomics. 2004. PMID: 15475259 - Molecular phylogeny of the antiangiogenic and neurotrophic serpin, pigment epithelium derived factor in vertebrates.
Xu X, Zhang SS, Barnstable CJ, Tombran-Tink J. Xu X, et al. BMC Genomics. 2006 Oct 4;7:248. doi: 10.1186/1471-2164-7-248. BMC Genomics. 2006. PMID: 17020603 Free PMC article. - Raising the estimate of functional human sequences.
Pheasant M, Mattick JS. Pheasant M, et al. Genome Res. 2007 Sep;17(9):1245-53. doi: 10.1101/gr.6406307. Epub 2007 Aug 9. Genome Res. 2007. PMID: 17690206 Review. - Retrotransposal integration of mobile genetic elements in human diseases.
Miki Y. Miki Y. J Hum Genet. 1998;43(2):77-84. doi: 10.1007/s100380050045. J Hum Genet. 1998. PMID: 9621510 Review.
Cited by
- Massively Parallel Functional Analysis of BRCA1 RING Domain Variants.
Starita LM, Young DL, Islam M, Kitzman JO, Gullingsrud J, Hause RJ, Fowler DM, Parvin JD, Shendure J, Fields S. Starita LM, et al. Genetics. 2015 Jun;200(2):413-22. doi: 10.1534/genetics.115.175802. Epub 2015 Mar 30. Genetics. 2015. PMID: 25823446 Free PMC article. - Exome sequencing and genome-wide linkage analysis in 17 families illustrate the complex contribution of TTN truncating variants to dilated cardiomyopathy.
Norton N, Li D, Rampersaud E, Morales A, Martin ER, Zuchner S, Guo S, Gonzalez M, Hedges DJ, Robertson PD, Krumm N, Nickerson DA, Hershberger RE; National Heart, Lung, and Blood Institute GO Exome Sequencing Project and the Exome Sequencing Project Family Studies Project Team. Norton N, et al. Circ Cardiovasc Genet. 2013 Apr;6(2):144-53. doi: 10.1161/CIRCGENETICS.111.000062. Epub 2013 Feb 15. Circ Cardiovasc Genet. 2013. PMID: 23418287 Free PMC article. - Functional dissection of human cardiac enhancers and noncoding de novo variants in congenital heart disease.
Xiao F, Zhang X, Morton SU, Kim SW, Fan Y, Gorham JM, Zhang H, Berkson PJ, Mazumdar N, Cao Y, Chen J, Hagen J, Liu X, Zhou P, Richter F, Shen Y, Ward T, Gelb BD, Seidman JG, Seidman CE, Pu WT. Xiao F, et al. Nat Genet. 2024 Mar;56(3):420-430. doi: 10.1038/s41588-024-01669-y. Epub 2024 Feb 20. Nat Genet. 2024. PMID: 38378865 Free PMC article. - Genomic analysis identified a potential novel molecular mechanism for high-altitude adaptation in sheep at the Himalayas.
Gorkhali NA, Dong K, Yang M, Song S, Kader A, Shrestha BS, He X, Zhao Q, Pu Y, Li X, Kijas J, Guan W, Han J, Jiang L, Ma Y. Gorkhali NA, et al. Sci Rep. 2016 Jul 22;6:29963. doi: 10.1038/srep29963. Sci Rep. 2016. PMID: 27444145 Free PMC article. - A further look at porcine chromosome 7 reveals VRTN variants associated with vertebral number in Chinese and Western pigs.
Fan Y, Xing Y, Zhang Z, Ai H, Ouyang Z, Ouyang J, Yang M, Li P, Chen Y, Gao J, Li L, Huang L, Ren J. Fan Y, et al. PLoS One. 2013 Apr 24;8(4):e62534. doi: 10.1371/journal.pone.0062534. Print 2013. PLoS One. 2013. PMID: 23638110 Free PMC article.
References
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301-1310. - PubMed
- Arnone, M.I. and Davidson, E.H. 1997. The hardwiring of development: Organization and function of genomic regulatory systems. Development 124: 1851-1864. - PubMed
- Bejerano, G., Pheasant, M., Makunin, I., Stephen, S., Kent, W.J., Mattick, J.S., and Haussler, D. 2004. Ultraconserved elements in the human genome. Science 304: 1321-1325. - PubMed
- Berman, B.P., Nibu, Y., Pfeiffer, B.D., Tomancak, P., Celniker, S.E., Levine, M., Rubin, G.M., and Eisen, M.B. 2002. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. 99: 757-762. - PMC - PubMed
Web site references
- http://blast.wustl.edu; WU-BLAST homepage.
- http://www.repeatmasker.org; RepeatMasker homepage.
- http://mendel.stanford.edu/sidowlab; Sidow Lab homepage.
- http://genome.ucsc.edu; UCSC Genome Browser homepage.
- http://www.nisc.nih.gov/data; NISC Comparative Sequencing Program homepage.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous