Llama antibodies against a lactococcal protein located at the tip of the phage tail prevent phage infection - PubMed (original) (raw)
Comparative Study
Llama antibodies against a lactococcal protein located at the tip of the phage tail prevent phage infection
Hans J W De Haard et al. J Bacteriol. 2005 Jul.
Abstract
Bacteriophage p2 belongs to the most prevalent lactococcal phage group (936) responsible for considerable losses in industrial production of cheese. Immunization of a llama with bacteriophage p2 led to higher titers of neutralizing heavy-chain antibodies (i.e., devoid of light chains) than of the classical type of immunoglobulins. A panel of p2-specific single-domain antibody fragments was obtained using phage display technology, from which a group of potent neutralizing antibodies were identified. The antigen bound by these antibodies was identified as a protein with a molecular mass of 30 kDa, homologous to open reading frame 18 (ORF18) of phage sk1, another 936-like phage for which the complete genomic sequence is available. By the use of immunoelectron microscopy, the protein is located at the tip of the tail of the phage particle. The addition of purified ORF18 protein to a bacterial culture suppressed phage infection. This result and the inhibition of cell lysis by anti-ORF18 protein antibodies support the conclusion that the ORF18 protein plays a crucial role in the interaction of bacteriophage p2 with the surface receptors of Lactococcus lactis.
Figures
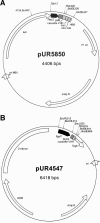
FIG. 1.
Plasmids used in this study. (A) Expression vector for large-scale production of VHH fragments in E. coli. PlacZ = IPTG-inducible promoter; pelB = signal sequence; myc and HIS6 encode tags; biot = biotinylation site. (B) Expression/integration vector used to transform S. cerevisiae and to express the various VHH fragments. Pgal7 = galactose-inducible promoter, SUC2 = invertase signal sequence, Tpgk = the PGK terminator. LEU2 is the LEU2 gene behind a defective promoter, which ensures, together with the 2 micron sequence, the multicopy integration of this vector on the rDNA locus.
FIG. 2.
Amino acid sequences of nonneutralizing (A) and neutralizing (B) bacteriophage-specific VHH fragments. Residues are numbered in accordance with Kabat et al. (23). The primer-encoded hinge region (long hinge) is also shown.
FIG. 3.
Acidification profiles and growth kinetics of phage-infected cultures as a function of the concentration of VHH#5. (A) Determination of the minimal concentration of VHH to neutralize an infection of 103 PFU/ml phage p2 by measuring pH and OD600. (B) Growth kinetics determined with OD600 for a culture containing 105 PFU/ml phage p2 and variable amounts of VHH.
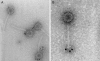
FIG. 4.
Immunoelectron microscopy with heavy-chain antibody fragments and 10-nm gold-labeled detection antibodies on bacteriophage p2. (A) Phage detected with nonneutralizing VHH#2. (B) Phage recognized by neutralizing fragment VHH#5.
FIG. 5.
Epitope mapping of anti-msp antibody fragment VHH#2. (A) Inserts of the VHH#2-recognized lambda gt11 clones (coded #1 to #6), which encode a shared segment of the carboxy-terminal region of the major structural protein. Numbering has been done as in the deposited sk1 genomic sequence (GenBank accession number AF011378). The earlier observed differences (25) between the shown p2-derived sequence and the sk1 sequence are indicated with boxes. (B) Pepscan results obtained with overlapping 15-mer peptides based on the shared sequence segment of the lambda gt11 clones. The peak corresponds with the peptide having the sequence NGQLAPGVYIVTFSA and therefore represents the epitope of VHH#2.
FIG. 5.
Epitope mapping of anti-msp antibody fragment VHH#2. (A) Inserts of the VHH#2-recognized lambda gt11 clones (coded #1 to #6), which encode a shared segment of the carboxy-terminal region of the major structural protein. Numbering has been done as in the deposited sk1 genomic sequence (GenBank accession number AF011378). The earlier observed differences (25) between the shown p2-derived sequence and the sk1 sequence are indicated with boxes. (B) Pepscan results obtained with overlapping 15-mer peptides based on the shared sequence segment of the lambda gt11 clones. The peak corresponds with the peptide having the sequence NGQLAPGVYIVTFSA and therefore represents the epitope of VHH#2.
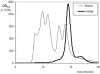
FIG. 6.
Size exclusion chromatography of VHH#5. VHH#5 and a mixture of marker proteins were analyzed on a Superdex 75 column, and the measured OD214 was plotted against the retention time. The peaks of the marker proteins (13.7, 25, 43, 67, and 2,000 kDa) are shown.
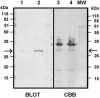
FIG. 7.
Specificity of neutralizing VHH#5 determined by Western blot analysis. Phage sk1 was loaded (lane 1, 6 × 109 PFU; lane 2, 3 × 1010 PFU) onto a 15% gel, blotted, and incubated with VHH#5. The other part of the gel (lane 3, 1 × 1010 PFU; lane 4, 2 × 1010 PFU) was stained with Coomassie brilliant blue (CBB). The experimentally determined amino-terminal sequence (ThrIleLysAsnPheThrPhePheSerProAsnSerThrGluPhe; the expected methionine as start residue has been clipped off) of the recognized protein (arrows) corresponds to the hypothetical gene product of ORF18. MW, molecular mass.
FIG. 8.
Western blot analysis with phage p2-derived proteins expressed in E. coli or complete bacteriophage particles of sk1. The antibody fragments VHH#2 and VHH#5 and postimmune llama serum (llama pAB blot) were used for detection. Purified ORF18, msp, lysin (lys), and a bacterial extract of E. coli cells containing the expression vector used (−) and phage sk1 (Φ) were loaded. The hexahistidine-tagged proteins were detected with anti-His monoclonal antibody (α-His).
FIG. 9.
Effect of ORF18 protein on infection as measured by acidification and the kinetics of growth. (A) Two concentrations of ORF18 were included in cultures of L. lactis C2 containing 102 PFU/ml bacteriophage p2. Infected and noninfected cultures were used as controls, while purified msp was added to another infected culture. (B) Partial protection by addition of ORF18 against higher phage titers (103 PFU/ml) as measured by the kinetics of growth (graph on the left). The dose-dependent neutralizing effect of ORF18 on cultures with phage titers of 102 PFU/ml was studied in more detail with a new batch of purified ORF18 (growth curves on the right).
FIG. 10.
Alignment of ORF18-encoded amino acid sequences of 936 bacteriophages p2, sk1, and bIL170. FASTA analysis revealed homology with the baseplate protein from temperate phage TP901-1 (residues 53 to 163).
Similar articles
- Identification of the receptor-binding protein in 936-species lactococcal bacteriophages.
Dupont K, Vogensen FK, Neve H, Bresciani J, Josephsen J. Dupont K, et al. Appl Environ Microbiol. 2004 Oct;70(10):5818-24. doi: 10.1128/AEM.70.10.5818-5824.2004. Appl Environ Microbiol. 2004. PMID: 15466519 Free PMC article. - Lactococcal bacteriophage p2 receptor-binding protein structure suggests a common ancestor gene with bacterial and mammalian viruses.
Spinelli S, Desmyter A, Verrips CT, de Haard HJ, Moineau S, Cambillau C. Spinelli S, et al. Nat Struct Mol Biol. 2006 Jan;13(1):85-9. doi: 10.1038/nsmb1029. Epub 2005 Dec 4. Nat Struct Mol Biol. 2006. PMID: 16327804 - Preventing phage lysis of Lactococcus lactis in cheese production using a neutralizing heavy-chain antibody fragment from llama.
Ledeboer AM, Bezemer S, de Hiaard JJ, Schaffers IM, Verrips CT, van Vliet C, Düsterhöft EM, Zoon P, Moineau S, Frenken LG. Ledeboer AM, et al. J Dairy Sci. 2002 Jun;85(6):1376-82. doi: 10.3168/jds.s0022-0302(02)74204-5. J Dairy Sci. 2002. PMID: 12146467 - Receptor-binding protein of Lactococcus lactis phages: identification and characterization of the saccharide receptor-binding site.
Tremblay DM, Tegoni M, Spinelli S, Campanacci V, Blangy S, Huyghe C, Desmyter A, Labrie S, Moineau S, Cambillau C. Tremblay DM, et al. J Bacteriol. 2006 Apr;188(7):2400-10. doi: 10.1128/JB.188.7.2400-2410.2006. J Bacteriol. 2006. PMID: 16547026 Free PMC article. - Flavour peptides: the potential role of Lactococcal peptidases in their production.
Mulholland F. Mulholland F. Biochem Soc Trans. 1991 Aug;19(3):685-90. doi: 10.1042/bst0190685. Biochem Soc Trans. 1991. PMID: 1783198 Review. No abstract available.
Cited by
- Biotechnological applications of recombinant single-domain antibody fragments.
de Marco A. de Marco A. Microb Cell Fact. 2011 Jun 9;10:44. doi: 10.1186/1475-2859-10-44. Microb Cell Fact. 2011. PMID: 21658216 Free PMC article. Review. - Viral infection modulation and neutralization by camelid nanobodies.
Desmyter A, Farenc C, Mahony J, Spinelli S, Bebeacua C, Blangy S, Veesler D, van Sinderen D, Cambillau C. Desmyter A, et al. Proc Natl Acad Sci U S A. 2013 Apr 9;110(15):E1371-9. doi: 10.1073/pnas.1301336110. Epub 2013 Mar 25. Proc Natl Acad Sci U S A. 2013. PMID: 23530214 Free PMC article. - Neutralisation of HIV-1 cell-cell spread by human and llama antibodies.
McCoy LE, Groppelli E, Blanchetot C, de Haard H, Verrips T, Rutten L, Weiss RA, Jolly C. McCoy LE, et al. Retrovirology. 2014 Oct 2;11:83. doi: 10.1186/s12977-014-0083-y. Retrovirology. 2014. PMID: 25700025 Free PMC article. - Structure and molecular assignment of lactococcal phage TP901-1 baseplate.
Bebeacua C, Bron P, Lai L, Vegge CS, Brøndsted L, Spinelli S, Campanacci V, Veesler D, van Heel M, Cambillau C. Bebeacua C, et al. J Biol Chem. 2010 Dec 10;285(50):39079-86. doi: 10.1074/jbc.M110.175646. Epub 2010 Oct 11. J Biol Chem. 2010. PMID: 20937834 Free PMC article. - Crystal structure of the receptor-binding protein head domain from Lactococcus lactis phage bIL170.
Ricagno S, Campanacci V, Blangy S, Spinelli S, Tremblay D, Moineau S, Tegoni M, Cambillau C. Ricagno S, et al. J Virol. 2006 Sep;80(18):9331-5. doi: 10.1128/JVI.01160-06. J Virol. 2006. PMID: 16940545 Free PMC article.
References
- Bissonnette, F., S. Labrie, H. Deveau, M. Lamoureux, and S. Moineau. 2000. Characterization of mesophilic mixed starter cultures used for the manufacture of aged cheddar cheese. J. Dairy Sci. 83:620-627. - PubMed
- Chandry, P. S., S. C. Moore, J. D. Boyce, B. E. Davidson, and A. J. Hillier. 1997. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol. Microbiol. 26:49-64. - PubMed
- Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources