Palm is expressed in both developing and adult mouse lens and retina - PubMed (original) (raw)
Palm is expressed in both developing and adult mouse lens and retina
Meryl Castellini et al. BMC Ophthalmol. 2005.
Abstract
Background: Paralemmin (Palm) is a prenyl-palmitoyl anchored membrane protein that can drive membrane and process formation in neurons. Earlier studies have shown brain preferred Palm expression, although this protein is a major water insoluble protein in chicken lens fiber cells and the Palm gene may be regulated by Pax6.
Methods: The expression profile of Palm protein in the embryonic, newborn and adult mouse eye as well as dissociated retinal neurons was determined by confocal immunofluorescence. The relative mRNA levels of Palm, Palmdelphin (PalmD) and paralemmin2 (Palm2) in the lens and retina were determined by real time rt-PCR.
Results: In the lens, Palm is already expressed at 9.5 dpc in the lens placode, and this expression is maintained in the lens vesicle throughout the formation of the adult lens. Palm is largely absent from the optic vesicle but is detectable at 10.5 dpc in the optic cup. In the developing retina, Palm expression transiently upregulates during the formation of optic nerve as well as in the formation of both the inner and outer plexiform layers. In short term dissociated chick retinal cultures, Palm protein is easily detectable, but the levels appear to reduce sharply as the cultures age. Palm mRNA was found at much higher levels relative to Palm2 or PalmD in both the retina and lens.
Conclusion: Palm is the major paralemmin family member expressed in the retina and lens and its expression in the retina transiently upregulates during active neurite outgrowth. The expression pattern of Palm in the eye is consistent with it being a Pax6 responsive gene. Since Palm is known to be able to drive membrane formation in brain neurons, it is possible that this molecule is crucial for the increase in membrane formation during lens fiber cell differentiation.
Figures
Figure 1
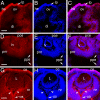
Localization of Palm protein during early mouse eye development. A-C, 9.5 dpc; D-F, 10.5 dpc; G-I, 12.5 dpc; A,D,G Palm; B,E,H cell nuclei stained with ToPro3, C,F,I, merge; Abbreviations- lp, lens placode; ov, optic vesicle; he, head ectoderm; di- lumen of the diencephalon; pce- presumptive corneal epithelium; lv- lens vesicle; pnr- presumptive neural retina; ppe- presumptive retinal pigmented epithelium; L- lens; nr- neural retina; ce- corneal epithelium; os- optic stalk. Arrowheads denote staining in developing neuronal processes that will grow through optic stalk to form the optic nerve. All scale bars are 77 μm. red- Palm; blue-ToPro3 DNA stain.
Figure 2
Localization of paralemmin protein in the mouse lens A-C 14.5 dpc; D 14.5 dpc negative control E-G One week post natal; H- 25 weeks postnatal. I- 25 weeks postnatal negative control A,E- paralemmin; B,F- cell nuclei stained with ToPro3; C, D, G, H, I- merge; Scale bars- A-C, 154 μm; D-G, 77 μm; red- paralemmin; blue-ToPro3 DNA stain.
Figure 3
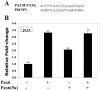
Pax6 proteins activate expression from a reporter consisting of four copies of a PAX6-binding site found in the putative 5' flanking sequence of the human PALM gene cloned upstream of the E4 basal promoter. (A) An alignment between the PAX6 site found in the PALM gene and a consensus paired domain Pax6-binding site, P6CON. Non-conserved nucleotides are shown in lower case letters. (B) Results of co-transfections in 293T cells. 200 ng of Pax6 and 25 ng of Pax6(5a) expression plasmids were used as indicated per experiment. The data were normalized using Renilla luciferase [31] and are expressed as a relative ratio of promoter activity in the presence of Pax6 compared to the presence of empty vector, pKW10.
Figure 4
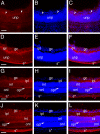
Localization of paralemmin protein during mouse retinal development A-C, 16.5 dpc, arrowheads- emerging inner plexiform layer; D-F 1 day pn; G-I 1 week pn; J-L 2 week pn; A,D,G,J- paralemmin; B,E,H,K- cell nuclei stained with ToPro3; C,F,I,L- merge; Abbreviations- unp- undifferentiated retinal precursors; gc- ganglion cell; ipl- inner plexiform layer; s*- background staining in the sclera; inl- inner nuclear layer; opl- outer plexiform layer; onl- outer nuclear layer. All scale bars are 77 μm. red- paralemmin; blue-ToPro3 DNA stain.
Figure 5
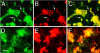
Expression and localization of Palm in chick retinal cultures. Cultures were immunostained with polyclonal anti-Palm (A, D) and RT-97 anti-neurofilament (B, E) antibodies after 2 (A-C) or 7 (D-F) days in culture. For each pair, the merged images are shown in C and F. After 2 days in culture, Palm is present on most cells at cell borders as well as intracellular puncta (A). Fine processes resembling axons (arrows) that are sometimes positive for RT-97 (B) are also labeled. After 7 days in culture, Palm staining appears punctate but more diffuse (D), and does not appear to be localized on the numerous long processes stained with RT-97 (F). Bar in D, 25μm. Green- Palm; Red-RT-97;
Figure 6
Relative levels of Palm, Palm2 and PalmD transcripts in the lens, cerebellum forebrain and retina. All data are expressed as a relative to the amount of B2M in the sample.
Similar articles
- Analysis of partner of inscuteable (mPins) expression in the developing mouse eye.
Raji B, Dansault A, Vieira V, de la Houssaye G, Lacassagne E, Kobetz A, Arbogast L, Dufier JL, Blumer JB, Menasche M, Abitbol M. Raji B, et al. Mol Vis. 2008;14:2575-96. Epub 2008 Dec 31. Mol Vis. 2008. PMID: 19122831 Free PMC article. - Expression of tissue plasminogen activator during eye development.
Collinge JE, Simirskii VN, Duncan MK. Collinge JE, et al. Exp Eye Res. 2005 Jul;81(1):90-6. doi: 10.1016/j.exer.2005.01.014. Exp Eye Res. 2005. PMID: 15978259 - Developmental expression of three small GTPases in the mouse eye.
Mitchell DC, Bryan BA, Liu JP, Liu WB, Zhang L, Qu J, Zhou X, Liu M, Li DW. Mitchell DC, et al. Mol Vis. 2007 Jul 13;13:1144-53. Mol Vis. 2007. PMID: 17653061 Free PMC article. - Molecular regulators involved in vertebrate eye development.
Jean D, Ewan K, Gruss P. Jean D, et al. Mech Dev. 1998 Aug;76(1-2):3-18. doi: 10.1016/s0925-4773(98)00117-8. Mech Dev. 1998. PMID: 9767078 Review. - The zebrafish eye: developmental and genetic analysis.
Easter SS Jr, Malicki JJ. Easter SS Jr, et al. Results Probl Cell Differ. 2002;40:346-70. doi: 10.1007/978-3-540-46041-1_17. Results Probl Cell Differ. 2002. PMID: 12353485 Review.
Cited by
- Tight binding of proteins to membranes from older human cells.
Truscott RJ, Comte-Walters S, Ablonczy Z, Schwacke JH, Berry Y, Korlimbinis A, Friedrich MG, Schey KL. Truscott RJ, et al. Age (Dordr). 2011 Dec;33(4):543-54. doi: 10.1007/s11357-010-9198-9. Epub 2010 Dec 23. Age (Dordr). 2011. PMID: 21181282 Free PMC article. - Cellular and subcellular localization of paralemmin-1, a protein involved in cell shape control, in the rat brain, adrenal gland and kidney.
Kutzleb C, Petrasch-Parwez E, Kilimann MW. Kutzleb C, et al. Histochem Cell Biol. 2007 Jan;127(1):13-30. doi: 10.1007/s00418-006-0209-y. Epub 2006 Jul 18. Histochem Cell Biol. 2007. PMID: 16847661 - MS/MS in silico subtraction-based proteomic profiling as an approach to facilitate disease gene discovery: application to lens development and cataract.
Aryal S, Anand D, Hernandez FG, Weatherbee BAT, Huang H, Reddy AP, Wilmarth PA, David LL, Lachke SA. Aryal S, et al. Hum Genet. 2020 Feb;139(2):151-184. doi: 10.1007/s00439-019-02095-5. Epub 2019 Dec 3. Hum Genet. 2020. PMID: 31797049 Free PMC article. - Genome-wide association studies identify two novel loci conferring susceptibility to diabetic retinopathy in Japanese patients with type 2 diabetes.
Imamura M, Takahashi A, Matsunami M, Horikoshi M, Iwata M, Araki SI, Toyoda M, Susarla G, Ahn J, Park KH, Kong J, Moon S, Sobrin L; International Diabetic Retinopathy and Genetics CONsortium (iDRAGON); Yamauchi T, Tobe K, Maegawa H, Kadowaki T, Maeda S. Imamura M, et al. Hum Mol Genet. 2021 May 17;30(8):716-726. doi: 10.1093/hmg/ddab044. Hum Mol Genet. 2021. PMID: 33607655 Free PMC article. - The membrane proteome of the mouse lens fiber cell.
Bassnett S, Wilmarth PA, David LL. Bassnett S, et al. Mol Vis. 2009 Nov 24;15:2448-63. Mol Vis. 2009. PMID: 19956408 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 EY012200/EY/NEI NIH HHS/United States
- EY015279/EY/NEI NIH HHS/United States
- P20 RR016472/RR/NCRR NIH HHS/United States
- EY012221/EY/NEI NIH HHS/United States
- NS40317/NS/NINDS NIH HHS/United States
- R01 EY012221/EY/NEI NIH HHS/United States
- EY12200/EY/NEI NIH HHS/United States
- P20 RR16472/RR/NCRR NIH HHS/United States
- R01 EY015279/EY/NEI NIH HHS/United States
- R21 NS040317/NS/NINDS NIH HHS/United States
- EY14237/EY/NEI NIH HHS/United States
- R01 EY014237/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases