TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing - PubMed (original) (raw)
TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing
Thimmaiah P Chendrimada et al. Nature. 2005.
Abstract
MicroRNAs (miRNAs) are generated by a two-step processing pathway to yield RNA molecules of approximately 22 nucleotides that negatively regulate target gene expression at the post-transcriptional level. Primary miRNAs are processed to precursor miRNAs (pre-miRNAs) by the Microprocessor complex. These pre-miRNAs are cleaved by the RNase III Dicer to generate mature miRNAs that direct the RNA-induced silencing complex (RISC) to messenger RNAs with complementary sequence. Here we show that TRBP (the human immunodeficiency virus transactivating response RNA-binding protein), which contains three double-stranded, RNA-binding domains, is an integral component of a Dicer-containing complex. Biochemical analysis of TRBP-containing complexes revealed the association of Dicer-TRBP with Argonaute 2 (Ago2), the catalytic engine of RISC. The physical association of Dicer-TRBP and Ago2 was confirmed after the isolation of the ternary complex using Flag-tagged Ago2 cell lines. In vitro reconstitution assays demonstrated that TRBP is required for the recruitment of Ago2 to the small interfering RNA (siRNA) bound by Dicer. Knockdown of TRBP results in destabilization of Dicer and a consequent loss of miRNA biogenesis. Finally, depletion of the Dicer-TRBP complex via exogenously introduced siRNAs diminished RISC-mediated reporter gene silencing. These results support a role of the Dicer-TRBP complex not only in miRNA processing but also as a platform for RISC assembly.
Figures
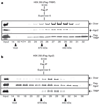
Figure 1. Isolation of a Dicer–TRBP-containing complex
a, Flag-Dicer isolated from a HEK293-derived cytoplasmic extract (S100) resolved by SDS–PAGE and visualized by silver staining (left panel), and western blot analysis using anti-Flag antibodies (right panel). Arrows point to the bands containing Dicer and TRBP proteins. The molecular masses of marker proteins (kDa) are indicated (right). Asterisks denote contaminating polypeptides that were also present in control immunoprecipitation (IP) from naive cells (Mock). b, Diagrammatic representation of the human TRBP protein and alignment with orthologous proteins. Conserved dsRBDs are depicted as black squares. Protein accession numbers (NCBI) are: AAB50581 (human TRBP), NP_003681 (human PRKRA), NP_03345 (mouse PRBP), AAH55390 (Danio rerio), MGC82499 (Xenopus), NP_609646 (Drosophila). aa, amino acids. c, The Dicer–TRBP complex isolated from S100 by Flag-TRBP, fractionated by Superose 6 gel filtration, and visualized by silver staining and western blot analysis. INP denotes the input to the column; asterisks denote the common contaminants (SKB1 and MEP50). Fractions of the column are shown along the top; molecular mass markers are indicated along the bottom. d, Silver stains of recombinant Dicer (rDicer) and recombinant Dicer–TRBP complex (rDicer + rTRBP) reconstituted and purified by gel filtration. e, Silver stain analysis of recombinant TRBP fractionated by gel filtration.
Figure 2. The Dicer–TRBP complex processes miRNAs and associates with siRNA to form a ternary complex with Ago2
a, Pre-miRNA processing assay (left panel). The pre-miRNAs generated by the Microprocessor complex were subjected to a second step of processing using recombinant Dicer, recombinant Dicer–TRBP, or fraction 28 of the Superose 6 column of immunopurified TRBP complex (Flag-TRBP) to yield a 22-nucleotide mature miRNA. The right panel shows the long double-stranded RNA processing assay. Double-stranded RNA was incubated with recombinant Dicer or recombinant Dicer–TRBP to yield a 22-nucleotide mature miRNA. b, EMSA performed using radiolabelled double-stranded siRNA. Recombinant Dicer and recombinant Dicer–TRBP (fraction 28 of the Superose 6 column) was analysed for complex formation with siRNA. 1× corresponds to 20 ng of Dicer and 10 ng of recombinant Ago2. c, EMSA performed using radiolabelled siRNA. Recombinant Dicer–TRBP (fraction 28 of the Superose 6 column) and recombinant TRBP (fraction 32 of the Superose 6 column) were analysed for complex formation with siRNA. 1× corresponds to 20 ng for each protein. d, EMSA performed with Dicer–TRBP (fraction 28 of the Superose 6 column corresponding to Flag-TRBP) and addition of recombinant Ago2. 1 × corresponds to 5 ng of native complex and 2.5 ng of recombinant Ago2. Arrows indicate the two ribonucleoprotein complexes.
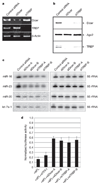
Figure 3. Stable association of Dicer–TRBP with Ago2
a, Western blot analysis of Flag-TRBP affinity eluate fractionated by Superose 6 gel filtration. Column fractions were analysed using the antibodies indicated. b, Western blot analysis of Flag-Ago2 affinity eluate fractionated by Superose 6 gel filtration. Column fractions were analysed using the antibodies indicated.
Figure 4. The Dicer–TRBP complex is required for miRNA biogenesis and post-transcriptional gene silencing
a, Analysis of the transcript levels of TRBP, Dicer and β-actin by RT–PCR, after treatment of HeLa cells with a representative siRNA to TFII-I (control), TRBP and Dicer. b, Western blot analysis of Flag-Ago2 immunoprecipitate after siRNA-mediated knockdown of TFII-I (control), TRBP and Dicer. c, Northern blot analysis of miR-16, miR-23, let-7a-1 and miR-20 after treatment of HeLa cells with the indicated siRNA. d, HeLa cells co-transfected with firefly and Renilla luciferase plasmids and a combination of siRNA targeting firefly luciferase and one of the siRNAs shown in the bottom of the figure. Firefly luciferase activity was normalized relative to that of Renilla luciferase. Error bars represent standard error of the mean for three independent experiments.
Similar articles
- The role of PACT in the RNA silencing pathway.
Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. Lee Y, et al. EMBO J. 2006 Feb 8;25(3):522-32. doi: 10.1038/sj.emboj.7600942. Epub 2006 Jan 19. EMBO J. 2006. PMID: 16424907 Free PMC article. - A human, ATP-independent, RISC assembly machine fueled by pre-miRNA.
Maniataki E, Mourelatos Z. Maniataki E, et al. Genes Dev. 2005 Dec 15;19(24):2979-90. doi: 10.1101/gad.1384005. Genes Dev. 2005. PMID: 16357216 Free PMC article. - TRBP ensures efficient Dicer processing of precursor microRNA in RNA-crowded environments.
Fareh M, Yeom KH, Haagsma AC, Chauhan S, Heo I, Joo C. Fareh M, et al. Nat Commun. 2016 Dec 9;7:13694. doi: 10.1038/ncomms13694. Nat Commun. 2016. PMID: 27934859 Free PMC article. - Intracellular localization and routing of miRNA and RNAi pathway components.
Ohrt T, Muetze J, Svoboda P, Schwille P. Ohrt T, et al. Curr Top Med Chem. 2012;12(2):79-88. doi: 10.2174/156802612798919132. Curr Top Med Chem. 2012. PMID: 22196276 Review. - MicroRNA Expression: Protein Participants in MicroRNA Regulation.
King VM, Borchert GM. King VM, et al. Methods Mol Biol. 2017;1617:27-37. doi: 10.1007/978-1-4939-7046-9_2. Methods Mol Biol. 2017. PMID: 28540674 Review.
Cited by
- LncRNA HBL1 is required for genome-wide PRC2 occupancy and function in cardiogenesis from human pluripotent stem cells.
Liu J, Liu S, Han L, Sheng Y, Zhang Y, Kim IM, Wan J, Yang L. Liu J, et al. Development. 2021 Jul 1;148(13):dev199628. doi: 10.1242/dev.199628. Epub 2021 Jun 28. Development. 2021. PMID: 34027990 Free PMC article. - Circulating miRNAs: cell-cell communication function?
Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B. Turchinovich A, et al. Front Genet. 2013 Jun 28;4:119. doi: 10.3389/fgene.2013.00119. Print 2013. Front Genet. 2013. PMID: 23825476 Free PMC article. - Characterization of a naturally occurring truncated Dicer.
Mosca N, Starega-Roslan J, Castiello F, Russo A, Krzyzosiak WJ, Potenza N. Mosca N, et al. Mol Biol Rep. 2015 Aug;42(8):1333-40. doi: 10.1007/s11033-015-3878-6. Epub 2015 Apr 25. Mol Biol Rep. 2015. PMID: 25911188 - Quantitative functions of Argonaute proteins in mammalian development.
Wang D, Zhang Z, O'Loughlin E, Lee T, Houel S, O'Carroll D, Tarakhovsky A, Ahn NG, Yi R. Wang D, et al. Genes Dev. 2012 Apr 1;26(7):693-704. doi: 10.1101/gad.182758.111. Genes Dev. 2012. PMID: 22474261 Free PMC article. - The TAR-RNA binding protein is required for immunoresponses triggered by Cardiovirus infection.
Komuro A, Homma Y, Negoro T, Barber GN, Horvath CM. Komuro A, et al. Biochem Biophys Res Commun. 2016 Nov 11;480(2):187-193. doi: 10.1016/j.bbrc.2016.10.023. Epub 2016 Oct 13. Biochem Biophys Res Commun. 2016. PMID: 27743889 Free PMC article.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
- Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. - PubMed
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. - PubMed
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM040536/GM/NIGMS NIH HHS/United States
- R01 HL070045/HL/NHLBI NIH HHS/United States
- R01 GM040536-15/GM/NIGMS NIH HHS/United States
- P01 CA072765/CA/NCI NIH HHS/United States
- P01 CA072765-050002/CA/NCI NIH HHS/United States
- R01 HL070045-03/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases