POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end - PubMed (original) (raw)
POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end
Dirk Hockemeyer et al. EMBO J. 2005.
Abstract
The hallmarks of telomere dysfunction in mammals are reduced telomeric 3' overhangs, telomere fusions, and cell cycle arrest due to a DNA damage response. Here, we report on the phenotypes of RNAi-mediated inhibition of POT1, the single-stranded telomeric DNA-binding protein. A 10-fold reduction in POT1 protein in tumor cells induced neither telomere fusions nor cell cycle arrest. However, the 3' overhang DNA was reduced and all telomeres elicited a transient DNA damage response in G1, indicating that extensive telomere damage can occur without cell cycle arrest or telomere fusions. RNAi to POT1 also revealed its role in generating the correct sequence at chromosome ends. The recessed 5' end of the telomere, which normally ends on the sequence ATC-5', was changed to a random position within the AATCCC repeat. Thus, POT1 determines the structure of the 3' and 5' ends of human chromosomes, and its inhibition generates a novel combination of telomere dysfunction phenotypes in which chromosome ends behave transiently as sites of DNA damage, yet remain protected from nonhomologous end-joining.
Figures
Figure 1
POT1 and POT1–55 and their depletion with RNAi. (A) Schematic of two POT1 mRNAs and the proteins they encode. The immunoblot to the right shows HTC75 cells infected with the pLPC vector or the same vector expressing the POT1–55 form from V4 mRNA. Dark fill: OB folds. Light fill: TPP1 interacting domain. (B) Quantitative immunoblot to determine the level of POT1 knockdown in HelaS3 cells. Serial dilutions of vector protein extract were compared to the indicated relative cell equivalents of ex18 and ex8a knockdown cell extracts.
Figure 2
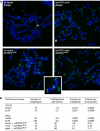
POT1 depletion does not induce significant levels of chromosome end fusions. (A) Metaphase spreads of the indicated HeLa cells with telomeric DNA detected by FISH (green). HeLa cells stably expressing the indicated shRNAs were infected with adenovirus expressing the TRF2 dominant-negative allele (AdTRF2ΔBΔM) or a control adenovirus (Adβgal), and processed for chromosome analysis after 2 days. M indicates a marker chromosome with internal TTAGGG repeats. The inset shows an enlarged image of one chromosome-type fusion from the AdTRF2ΔBΔM/POT1 shRNA panel. (B) Summary of the frequency of chromosome fusions in HeLa cells and HCT75 cells stably expressing POT1 shRNA and treated with the AdTRF2ΔBΔM adenovirus to inhibit TRF2. Chromosomes were analyzed 2 days after introduction of AdTRF2ΔBΔM and 15 days after knockdown of POT1.
Figure 3
Transient telomere damage response upon POT1 depletion. (A) TIFs induced by POT1 shRNA ex18. BJ/hTERT cells were processed for TIF analysis 8 days after infection and selection of the ex18 vector or the vector control by IF for TRF1 (red) and 53BP1 (green) or γ-H2AX (green). Merged images are shown with DAPI. (B) Quantification of the induction of TIFs by POT1 ex18 shRNA. IF for TRF1 and 53BP1 (see (A)) was used and randomly selected groups of cells were evaluated for the number of TRF1 signals per cell that contained a 53BP1 signal. The bars show the percentage of cells containing 10 or more TIFs. (C) The majority of the telomeres in TIF-positive cells colocalize with 53BP1. TIF-positive BJ (•) and BJ/hTERT (○) cells were selected and imaged using deconvolution software. Each point in the graph represents one TIF-positive cell and shows the number of TRF1 signals plotted versus the number of TRF1 signals containing 53BP1. Points above the line represent cells in which more than 50% of the TRF1 signals contained 53BP1. (D) Transient TIFs in G1. HeLa cells expressing POT1 shRNA ex18 or vector control were released from double-thymidine block and processed for IF at the indicated time points. FACS profiles are shown below the IF images. Top: 53BP1 signal (green) merged with DAPI (blue). Bottom: 53BP1, TRF1 (red), and DAPI signals merged. (E) Quantification of cell cycle dependence of TIF-positive cells. Quantification as in (B), using the cells shown in (D).
Figure 4
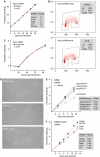
Differential effect of POT1 shRNA on proliferation of primary and transformed cells. (A) Graph showing growth curves of HeLa cells with and without POT1 shRNA. The inset shows the growth rate (PD/day) of these and additional HeLa cells with lowered POT1 level. (B) Graph showing proliferation of HeLa cells transfected with POT1 siRNA or control siRNA. (C) FACS profiles of BrdU-labeled HeLa cells infected with the indicated shRNA viruses. Insets show the % cells in G1, S, and G2/M. (D) Phase-contrast microscopic images of IMR90 cells 7 days after infection and selection with the indicated shRNA viruses stained for SA-β-galactosidase activity (Dimri et al, 1995) for 10 h. (E) Graph showing the effect of POT1 shRNA ex18 on the proliferation of IMR90 cells. The inset shows the growth rates (PD/day) in cells infected with the indicated shRNAs and a vector expressing a POT1 cDNA resistant to ex18 shRNA (POT1*). (F) Graph showing the effect of POT1 shRNAs on IMR90 cells transformed by introduction of SV40 large T antigen (pBabeNeoLT). The inset lists the growth rate of the cells.
Figure 5
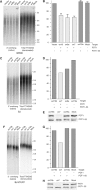
POT1 is required for the maintenance of the 3′ overhang. (A, C, F) In-gel assay for ss TTAGGG repeats with the indicated cell lines expressing the indicated shRNAs. DNA was isolated 7–10 days post-infection, cut with _Mbo_I and _Alu_I and processed by in-gel hybridization to a (CCCTAA)4 probe to detect ss TTAGGG repeats (left panels). The DNA was subsequently denatured in situ and rehybridized to the probe to detect the total TTAGGG repeat signal (right panels). (B, D, G) Bar graphs representing quantified overhang signals. Overhang signals in each lane were normalized to the total TTAGGG signal. The values are expressed relative to the value obtained with mock or vector (B) infected cells. The values in (B) represent averages of three experiments (SDs indicated). Below the bargraph, + and − indicate whether the shRNA targets POT1 and/or POT1–55. (E, H) Immunoblots of POT1 and POT1–55 levels in BJ (E) and BJ/hTERT (H) cells infected with retroviruses expressing the indicated shRNAs.
Figure 6
POT1 determines the sequence at the 5′ end of human chromosomes. (A) Schematic of the ends of human chromosomes and the 5′ telorette assay. The six telorettes and the 5′ ends to which they can ligate are shown. PCR primers used for amplification are shown schematically. (B–F) Products of the 5′ telorette assay using the indicated cell lines. Each telorette was used for 2–5 independent assays and the products were run in separate lanes. The sequences of the 5′ end detected with each telorette is shown above the groups of lanes. POT1* is a vector expressing full-length POT1 mutated to create resistance to the ex18 target site. The MWs of the detected products range from 1 to 8 kb.
Similar articles
- Coordinated interactions of multiple POT1-TPP1 proteins with telomere DNA.
Corriveau M, Mullins MR, Baus D, Harris ME, Taylor DJ. Corriveau M, et al. J Biol Chem. 2013 Jun 7;288(23):16361-16370. doi: 10.1074/jbc.M113.471896. Epub 2013 Apr 24. J Biol Chem. 2013. PMID: 23616058 Free PMC article. - In vivo stoichiometry of shelterin components.
Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. Takai KK, et al. J Biol Chem. 2010 Jan 8;285(2):1457-67. doi: 10.1074/jbc.M109.038026. Epub 2009 Oct 28. J Biol Chem. 2010. PMID: 19864690 Free PMC article. - TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase.
Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. Xin H, et al. Nature. 2007 Feb 1;445(7127):559-62. doi: 10.1038/nature05469. Epub 2007 Jan 21. Nature. 2007. PMID: 17237767 - POT1 mutations cause differential effects on telomere length leading to opposing disease phenotypes.
Zade NH, Khattar E. Zade NH, et al. J Cell Physiol. 2023 Jun;238(6):1237-1255. doi: 10.1002/jcp.31034. Epub 2023 May 14. J Cell Physiol. 2023. PMID: 37183325 Review. - Shelterin: the protein complex that shapes and safeguards human telomeres.
de Lange T. de Lange T. Genes Dev. 2005 Sep 15;19(18):2100-10. doi: 10.1101/gad.1346005. Genes Dev. 2005. PMID: 16166375 Review.
Cited by
- Cancer-associated TERT promoter mutations abrogate telomerase silencing.
Chiba K, Johnson JZ, Vogan JM, Wagner T, Boyle JM, Hockemeyer D. Chiba K, et al. Elife. 2015 Jul 21;4:e07918. doi: 10.7554/eLife.07918. Elife. 2015. PMID: 26194807 Free PMC article. - Telomeres and telomerase in cancer.
Artandi SE, DePinho RA. Artandi SE, et al. Carcinogenesis. 2010 Jan;31(1):9-18. doi: 10.1093/carcin/bgp268. Epub 2009 Nov 3. Carcinogenesis. 2010. PMID: 19887512 Free PMC article. Review. - Telomere Fragility and MiDAS: Managing the Gaps at the End of the Road.
Barnes RP, Thosar SA, Opresko PL. Barnes RP, et al. Genes (Basel). 2023 Jan 29;14(2):348. doi: 10.3390/genes14020348. Genes (Basel). 2023. PMID: 36833275 Free PMC article. Review. - Human POT1 protects the telomeric ds-ss DNA junction by capping the 5' end of the chromosome.
Tesmer VM, Brenner KA, Nandakumar J. Tesmer VM, et al. Science. 2023 Aug 18;381(6659):771-778. doi: 10.1126/science.adi2436. Epub 2023 Aug 17. Science. 2023. PMID: 37590346 Free PMC article. - Telomere dysfunction and cell survival: roles for distinct TIN2-containing complexes.
Kim SH, Davalos AR, Heo SJ, Rodier F, Zou Y, Beausejour C, Kaminker P, Yannone SM, Campisi J. Kim SH, et al. J Cell Biol. 2008 May 5;181(3):447-60. doi: 10.1083/jcb.200710028. Epub 2008 Apr 28. J Cell Biol. 2008. PMID: 18443218 Free PMC article.
References
- Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH (2001) Strand-specific postreplicative processing of mammalian telomeres. Science 293: 2462–2465 - PubMed
- Baird DM, Rowson J, Wynford-Thomas D, Kipling D (2003) Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat Genet 33: 203–207 - PubMed
- Baumann P, Cech TR (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AG01228/AG/NIA NIH HHS/United States
- AG16642/AG/NIA NIH HHS/United States
- R01 GM049046/GM/NIGMS NIH HHS/United States
- R01 AG001228/AG/NIA NIH HHS/United States
- R56 AG016642/AG/NIA NIH HHS/United States
- R37 GM049046/GM/NIGMS NIH HHS/United States
- R01 AG016642/AG/NIA NIH HHS/United States
- GM49046/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials