I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure - PubMed (original) (raw)
I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure
Emidio Capriotti et al. Nucleic Acids Res. 2005.
Abstract
I-Mutant2.0 is a support vector machine (SVM)-based tool for the automatic prediction of protein stability changes upon single point mutations. I-Mutant2.0 predictions are performed starting either from the protein structure or, more importantly, from the protein sequence. This latter task, to the best of our knowledge, is exploited for the first time. The method was trained and tested on a data set derived from ProTherm, which is presently the most comprehensive available database of thermodynamic experimental data of free energy changes of protein stability upon mutation under different conditions. I-Mutant2.0 can be used both as a classifier for predicting the sign of the protein stability change upon mutation and as a regression estimator for predicting the related DeltaDeltaG values. Acting as a classifier, I-Mutant2.0 correctly predicts (with a cross-validation procedure) 80% or 77% of the data set, depending on the usage of structural or sequence information, respectively. When predicting DeltaDeltaG values associated with mutations, the correlation of predicted with expected/experimental values is 0.71 (with a standard error of 1.30 kcal/mol) and 0.62 (with a standard error of 1.45 kcal/mol) when structural or sequence information are respectively adopted. Our web interface allows the selection of a predictive mode that depends on the availability of the protein structure and/or sequence. In this latter case, the web server requires only pasting of a protein sequence in a raw format. We therefore introduce I-Mutant2.0 as a unique and valuable helper for protein design, even when the protein structure is not yet known with atomic resolution.
Availability: http://gpcr.biocomp.unibo.it/cgi/predictors/I-Mutant2.0/I-Mutant2.0.cgi.
Figures
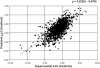
Figure 1
Correlation plot between the experimentally observed and the predicted ΔΔ_G_ values when the SVM method is structure based. The correlation is 0.71 and the corresponding standard error is 1.3 kcal/mol.
Figure 2
Snapshot of the I-Mutant2.0 input pages for the prediction of the protein stability change upon mutation with the protein structure through its PDB code (top) or starting from the sequence (bottom).
Similar articles
- Predicting protein stability changes from sequences using support vector machines.
Capriotti E, Fariselli P, Calabrese R, Casadio R. Capriotti E, et al. Bioinformatics. 2005 Sep 1;21 Suppl 2:ii54-8. doi: 10.1093/bioinformatics/bti1109. Bioinformatics. 2005. PMID: 16204125 - A neural-network-based method for predicting protein stability changes upon single point mutations.
Capriotti E, Fariselli P, Casadio R. Capriotti E, et al. Bioinformatics. 2004 Aug 4;20 Suppl 1:i63-8. doi: 10.1093/bioinformatics/bth928. Bioinformatics. 2004. PMID: 15262782 - CUPSAT: prediction of protein stability upon point mutations.
Parthiban V, Gromiha MM, Schomburg D. Parthiban V, et al. Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W239-42. doi: 10.1093/nar/gkl190. Nucleic Acids Res. 2006. PMID: 16845001 Free PMC article. - Prediction of protein stability upon point mutations.
Gromiha MM. Gromiha MM. Biochem Soc Trans. 2007 Dec;35(Pt 6):1569-73. doi: 10.1042/BST0351569. Biochem Soc Trans. 2007. PMID: 18031268 Review. - Sequence search methods and scoring functions for the design of protein structures.
Madaoui H, Becker E, Guerois R. Madaoui H, et al. Methods Mol Biol. 2006;340:183-206. doi: 10.1385/1-59745-116-9:183. Methods Mol Biol. 2006. PMID: 16957338 Review.
Cited by
- BALL-SNP: combining genetic and structural information to identify candidate non-synonymous single nucleotide polymorphisms.
Mueller SC, Backes C, Kalinina OV, Meder B, Stöckel D, Lenhof HP, Meese E, Keller A. Mueller SC, et al. Genome Med. 2015 Jul 1;7(1):65. doi: 10.1186/s13073-015-0190-y. eCollection 2015. Genome Med. 2015. PMID: 26191084 Free PMC article. - PROTS-RF: a robust model for predicting mutation-induced protein stability changes.
Li Y, Fang J. Li Y, et al. PLoS One. 2012;7(10):e47247. doi: 10.1371/journal.pone.0047247. Epub 2012 Oct 15. PLoS One. 2012. PMID: 23077576 Free PMC article. - Predicting folding free energy changes upon single point mutations.
Zhang Z, Wang L, Gao Y, Zhang J, Zhenirovskyy M, Alexov E. Zhang Z, et al. Bioinformatics. 2012 Mar 1;28(5):664-71. doi: 10.1093/bioinformatics/bts005. Epub 2012 Jan 11. Bioinformatics. 2012. PMID: 22238268 Free PMC article. - Acute Intermittent Porphyria: Predicted Pathogenicity of HMBS Variants Indicates Extremely Low Penetrance of the Autosomal Dominant Disease.
Chen B, Solis-Villa C, Hakenberg J, Qiao W, Srinivasan RR, Yasuda M, Balwani M, Doheny D, Peter I, Chen R, Desnick RJ. Chen B, et al. Hum Mutat. 2016 Nov;37(11):1215-1222. doi: 10.1002/humu.23067. Epub 2016 Sep 5. Hum Mutat. 2016. PMID: 27539938 Free PMC article. - A comprehensive in silico investigation into the pathogenic SNPs in the RTEL1 gene and their biological consequences.
Tanshee RR, Mahmud Z, Nabi AHMN, Sayem M. Tanshee RR, et al. PLoS One. 2024 Sep 6;19(9):e0309713. doi: 10.1371/journal.pone.0309713. eCollection 2024. PLoS One. 2024. PMID: 39240887 Free PMC article.
References
- Casadio R., Compiani M., Fariselli P., Vivarelli F. Predicting free energy contributions to the conformational stability of folded proteins from the residue sequence with radial basis function networks. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:81–88. - PubMed
- Gilis D., Rooman M. Predicting protein stability changes upon mutation using database-derived potentials: solvent accessibility determines the importance of local versus non-local interactions along the sequence. J. Mol. Biol. 1997;272:276–290. - PubMed
- Guerois R., Nielsen J.E., Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 2002;320:369–387. - PubMed
- Khatun J., Khare S.D., Dokholyan N.V. Can contact potentials reliably predict stability of proteins? J. Mol. Biol. 2004;336:1223–1238. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases