AntiHunter 2.0: increased speed and sensitivity in searching BLAST output for EST antisense transcripts - PubMed (original) (raw)
AntiHunter 2.0: increased speed and sensitivity in searching BLAST output for EST antisense transcripts
Giovanni Lavorgna et al. Nucleic Acids Res. 2005.
Abstract
An increasing number of eukaryotic and prokaryotic genes are being found to have natural antisense transcripts (NATs). There is also growing evidence to suggest that antisense transcription could play a key role in many human diseases. Consequently, there have been several recent attempts to set up computational procedures aimed at identifying novel NATs. Our group has developed the AntiHunter program for the identification of expressed sequence tag (EST) antisense transcripts from BLAST output. In order to perform an analysis, the program requires a genomic sequence plus an associated list of transcript names and coordinates of the genomic region. After masking the repeated regions, the program carries out a BLASTN search of this sequence in the selected EST database, reporting via email the EST entries that reveal an antisense transcript according to the user-supplied list. Here, we present the newly developed version 2.0 of the AntiHunter tool. Several improvements have been added to this version of the program in order to increase its ability to detect a larger number of antisense ESTs. As a result, AntiHunter can now detect, on average, >45% more antisense ESTs with little or no increase in the percentage of the false positives. We also raised the maximum query size to 3 Mb (previously 1 Mb). Moreover, we found that a reasonable trade-off between the program search sensitivity and the maximum allowed size of the input-query sequence could be obtained by querying the database with the MEGABLAST program, rather than by using the BLAST one. We now offer this new opportunity to users, i.e. if choosing the MEGABLAST option, users can input a query sequence up to 30 Mb long, thus considerably improving the possibility to analyze longer query regions. The AntiHunter tool is freely available at http://bioinfo.crs4.it/AH2.0.
Figures
Figure 1
Parameterizing the value of constant ‘Bases_Searched_For_Splicing_Consensi’ in AntiHunter. The constant ‘Bases_Searched_For_Splicing_Consensi’ determines the number of bases located upstream and downstream of the edge of a BLAST alignment between a genomic and an EST sequence that are searched for in the presence of splicing consensi. It used to be set to a fixed value of 5 in the AntiHunter program. This low value made unfeasible the detection of alignments like those shown in (A), where up to 11 spurious bases (shown in boldface uppercase) are added at the edge of the alignment between a query genomic sequence from MYCN locus (coordinates: chr2:16024168-16039977 from the release hg17 of the UCSC genome browser) and the EST AA609982. The specialized programs SIM4 (
) correctly detects the alignment boundaries of the alignment, as shown in (B). In AntiHunter 2.0, this hard-coded constant value has been parameterized, allowing the user to experiment with it: the splicing consensi are indeed correctly identified by AntiHunter when using a value >11.
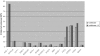
Figure 2
Benchmarking the performance of AntiHunter 2.0. The capability of AntiHunter 2.0 to detect EST antisense transcripts was compared with that of AntiHunter on a test case of 15 genomic regions, containing overlapping transcriptional units previously described in literature in mammalian genomes (for details see
). As a result, AntiHunter 2.0 detected a significantly larger number, 272 versus 186, of antisense ESTs than the previous version of the program. The newly detected ESTs belonged to six different genomic loci (ASE-1, RFPL3S, RFPL1, MYCN, FGF2 and THRA).
Similar articles
- AntiHunter: searching BLAST output for EST antisense transcripts.
Lavorgna G, Sessa L, Guffanti A, Lassandro L, Casari G. Lavorgna G, et al. Bioinformatics. 2004 Mar 1;20(4):583-5. doi: 10.1093/bioinformatics/btg460. Epub 2004 Jan 22. Bioinformatics. 2004. PMID: 14990456 - OrfPredictor: predicting protein-coding regions in EST-derived sequences.
Min XJ, Butler G, Storms R, Tsang A. Min XJ, et al. Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W677-80. doi: 10.1093/nar/gki394. Nucleic Acids Res. 2005. PMID: 15980561 Free PMC article. - TargetIdentifier: a webserver for identifying full-length cDNAs from EST sequences.
Min XJ, Butler G, Storms R, Tsang A. Min XJ, et al. Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W669-72. doi: 10.1093/nar/gki436. Nucleic Acids Res. 2005. PMID: 15980559 Free PMC article. - A hitchhiker's guide to expressed sequence tag (EST) analysis.
Nagaraj SH, Gasser RB, Ranganathan S. Nagaraj SH, et al. Brief Bioinform. 2007 Jan;8(1):6-21. doi: 10.1093/bib/bbl015. Epub 2006 May 23. Brief Bioinform. 2007. PMID: 16772268 Review. - In search of antisense.
Lavorgna G, Dahary D, Lehner B, Sorek R, Sanderson CM, Casari G. Lavorgna G, et al. Trends Biochem Sci. 2004 Feb;29(2):88-94. doi: 10.1016/j.tibs.2003.12.002. Trends Biochem Sci. 2004. PMID: 15102435 Review.
Cited by
- TRPM2: a candidate therapeutic target for treating neurological diseases.
Belrose JC, Jackson MF. Belrose JC, et al. Acta Pharmacol Sin. 2018 May;39(5):722-732. doi: 10.1038/aps.2018.31. Epub 2018 Apr 19. Acta Pharmacol Sin. 2018. PMID: 29671419 Free PMC article. Review. - Expression-profiling of apoptosis induced by ablation of the long ncRNA TRPM2-AS in prostate cancer cell.
Lavorgna G, Chiacchiera F, Briganti A, Montorsi F, Pasini D, Salonia A. Lavorgna G, et al. Genom Data. 2014 Nov 7;3:4-5. doi: 10.1016/j.gdata.2014.10.020. eCollection 2015 Mar. Genom Data. 2014. PMID: 26484139 Free PMC article. - Noncoding RNA synthesis and loss of Polycomb group repression accompanies the colinear activation of the human HOXA cluster.
Sessa L, Breiling A, Lavorgna G, Silvestri L, Casari G, Orlando V. Sessa L, et al. RNA. 2007 Feb;13(2):223-39. doi: 10.1261/rna.266707. Epub 2006 Dec 21. RNA. 2007. PMID: 17185360 Free PMC article. - Analysis of transcripts from predicted open reading frames of Musca domestica salivary gland hypertrophy virus.
Salem TZ, Garcia-Maruniak A, Lietze VU, Maruniak JE, Boucias DG. Salem TZ, et al. J Gen Virol. 2009 May;90(Pt 5):1270-1280. doi: 10.1099/vir.0.009613-0. Epub 2009 Mar 4. J Gen Virol. 2009. PMID: 19264592 Free PMC article.
References
- Wagner E.G., Altuvia S., Romby P. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 2002;46:361–398. - PubMed
- Sleutels F., Zwart R., Barlow D.P. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous