WebGestalt: an integrated system for exploring gene sets in various biological contexts - PubMed (original) (raw)
WebGestalt: an integrated system for exploring gene sets in various biological contexts
Bing Zhang et al. Nucleic Acids Res. 2005.
Abstract
High-throughput technologies have led to the rapid generation of large-scale datasets about genes and gene products. These technologies have also shifted our research focus from 'single genes' to 'gene sets'. We have developed a web-based integrated data mining system, WebGestalt (http://genereg.ornl.gov/webgestalt/), to help biologists in exploring large sets of genes. WebGestalt is composed of four modules: gene set management, information retrieval, organization/visualization, and statistics. The management module uploads, saves, retrieves and deletes gene sets, as well as performs Boolean operations to generate the unions, intersections or differences between different gene sets. The information retrieval module currently retrieves information for up to 20 attributes for all genes in a gene set. The organization/visualization module organizes and visualizes gene sets in various biological contexts, including Gene Ontology, tissue expression pattern, chromosome distribution, metabolic and signaling pathways, protein domain information and publications. The statistics module recommends and performs statistical tests to suggest biological areas that are important to a gene set and warrant further investigation. In order to demonstrate the use of WebGestalt, we have generated 48 gene sets with genes over-represented in various human tissue types. Exploration of all the 48 gene sets using WebGestalt is available for the public at http://genereg.ornl.gov/webgestalt/wg\_enrich.php.
Figures
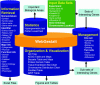
Figure 1
Schematic overview of WebGestalt. WebGestalt is composed of four main modules: gene set management, information retrieval, organization/visualization and statistics. The gene set management module uploads, saves, retrieves and deletes gene sets, as well as performs Boolean operations to generate the unions, intersections and differences between gene sets. The uploading tool accepts datasets defined by experiment data, GO categories or chromosome location ranges. WebGestalt is flexible in the input identifier (Entrez Gene ID, Swiss-Prot ID, Ensembl ID, Unigene ID, gene symbol and Affymetrix Probe Set ID). The saving tool saves sub-sets of genes generated by the organization/visualization module. The information retrieval module currently retrieves information for up to 20 attributes for all genes in a gene set, including nomenclatures, various gene identifiers, map and functional information. Retrieved information can be exported to Microsoft Excel files. The organization/visualization module organizes and visualizes a gene set in figures or tables using eight sub-modules: GO Tree, Tissue Expression Bar Chart, Chromosome Distribution Chart, KEGG Table and Maps, BioCarta Table and Maps, Protein Domain Table, PubMed Table and GRIF Table. The statistics module provides two statistical tests, the hypergeometric test and Fisher's exact test and suggests important biological areas in a gene set.
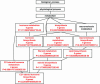
Figure 2
Enriched DAG under ‘biological process’ for a set of 23 genes that are significantly over-represented in adrenal cortex, using all genes in the human genome as a reference. The enriched GO categories are brought together and visualized as a DAG. Categories in red are enriched ones while those in black are non-enriched parents. Listed in the boxes are the name of the GO category, the number of genes in the category and the _P_-value indicating the significance of enrichment.
Similar articles
- GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies.
Zhang B, Schmoyer D, Kirov S, Snoddy J. Zhang B, et al. BMC Bioinformatics. 2004 Feb 18;5:16. doi: 10.1186/1471-2105-5-16. BMC Bioinformatics. 2004. PMID: 14975175 Free PMC article. - WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013.
Wang J, Duncan D, Shi Z, Zhang B. Wang J, et al. Nucleic Acids Res. 2013 Jul;41(Web Server issue):W77-83. doi: 10.1093/nar/gkt439. Epub 2013 May 23. Nucleic Acids Res. 2013. PMID: 23703215 Free PMC article. - WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit.
Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. Wang J, et al. Nucleic Acids Res. 2017 Jul 3;45(W1):W130-W137. doi: 10.1093/nar/gkx356. Nucleic Acids Res. 2017. PMID: 28472511 Free PMC article. - GeneKeyDB: a lightweight, gene-centric, relational database to support data mining environments.
Kirov SA, Peng X, Baker E, Schmoyer D, Zhang B, Snoddy J. Kirov SA, et al. BMC Bioinformatics. 2005 Mar 24;6:72. doi: 10.1186/1471-2105-6-72. BMC Bioinformatics. 2005. PMID: 15790402 Free PMC article. - Exploiting big biology: integrating large-scale biological data for function inference.
Marcotte E, Date S. Marcotte E, et al. Brief Bioinform. 2001 Dec;2(4):363-74. doi: 10.1093/bib/2.4.363. Brief Bioinform. 2001. PMID: 11808748 Review.
Cited by
- Ozone-derived oxysterols impair lung macrophage phagocytosis via adduction of some phagocytosis receptors.
Duffney PF, Kim HH, Porter NA, Jaspers I. Duffney PF, et al. J Biol Chem. 2020 Sep 4;295(36):12727-12738. doi: 10.1074/jbc.RA120.013699. Epub 2020 Jul 20. J Biol Chem. 2020. PMID: 32690608 Free PMC article. - The Effects of Ivermectin on Brugia malayi Females In Vitro: A Transcriptomic Approach.
Ballesteros C, Tritten L, O'Neill M, Burkman E, Zaky WI, Xia J, Moorhead A, Williams SA, Geary TG. Ballesteros C, et al. PLoS Negl Trop Dis. 2016 Aug 16;10(8):e0004929. doi: 10.1371/journal.pntd.0004929. eCollection 2016 Aug. PLoS Negl Trop Dis. 2016. PMID: 27529747 Free PMC article. - A genome-wide association study implicates the APOE locus in nonpathological cognitive ageing.
Davies G, Harris SE, Reynolds CA, Payton A, Knight HM, Liewald DC, Lopez LM, Luciano M, Gow AJ, Corley J, Henderson R, Murray C, Pattie A, Fox HC, Redmond P, Lutz MW, Chiba-Falek O, Linnertz C, Saith S, Haggarty P, McNeill G, Ke X, Ollier W, Horan M, Roses AD, Ponting CP, Porteous DJ, Tenesa A, Pickles A, Starr JM, Whalley LJ, Pedersen NL, Pendleton N, Visscher PM, Deary IJ. Davies G, et al. Mol Psychiatry. 2014 Jan;19(1):76-87. doi: 10.1038/mp.2012.159. Epub 2012 Dec 4. Mol Psychiatry. 2014. PMID: 23207651 Free PMC article. - Protection against COVID-19 injury by qingfei paidu decoction via anti-viral, anti-inflammatory activity and metabolic programming.
Chen J, Wang YK, Gao Y, Hu LS, Yang JW, Wang JR, Sun WJ, Liang ZQ, Cao YM, Cao YB. Chen J, et al. Biomed Pharmacother. 2020 Sep;129:110281. doi: 10.1016/j.biopha.2020.110281. Epub 2020 May 25. Biomed Pharmacother. 2020. PMID: 32554251 Free PMC article. - Transcriptome analysis of Inbred Long Sleep and Inbred Short Sleep mice.
Darlington TM, Ehringer MA, Larson C, Phang TL, Radcliffe RA. Darlington TM, et al. Genes Brain Behav. 2013 Mar;12(2):263-74. doi: 10.1111/gbb.12018. Genes Brain Behav. 2013. PMID: 23433184 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- P01 DA015027/DA/NIDA NIH HHS/United States
- R21 AA013532/AA/NIAAA NIH HHS/United States
- P01-DA015027/DA/NIDA NIH HHS/United States
- U01-AA013532/AA/NIAAA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical