Motifs, themes and thematic maps of an integrated Saccharomyces cerevisiae interaction network - PubMed (original) (raw)
Motifs, themes and thematic maps of an integrated Saccharomyces cerevisiae interaction network
Lan V Zhang et al. J Biol. 2005.
Abstract
Background: Large-scale studies have revealed networks of various biological interaction types, such as protein-protein interaction, genetic interaction, transcriptional regulation, sequence homology, and expression correlation. Recurring patterns of interconnection, or 'network motifs', have revealed biological insights for networks containing either one or two types of interaction.
Results: To study more complex relationships involving multiple biological interaction types, we assembled an integrated Saccharomyces cerevisiae network in which nodes represent genes (or their protein products) and differently colored links represent the aforementioned five biological interaction types. We examined three- and four-node interconnection patterns containing multiple interaction types and found many enriched multi-color network motifs. Furthermore, we showed that most of the motifs form 'network themes' -- classes of higher-order recurring interconnection patterns that encompass multiple occurrences of network motifs. Network themes can be tied to specific biological phenomena and may represent more fundamental network design principles. Examples of network themes include a pair of protein complexes with many inter-complex genetic interactions -- the 'compensatory complexes' theme. Thematic maps -- networks rendered in terms of such themes -- can simplify an otherwise confusing tangle of biological relationships. We show this by mapping the S. cerevisiae network in terms of two specific network themes.
Conclusion: Significantly enriched motifs in an integrated S. cerevisiae interaction network are often signatures of network themes, higher-order network structures that correspond to biological phenomena. Representing networks in terms of network themes provides a useful simplification of complex biological relationships.
Figures
Figure 1
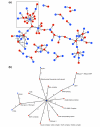
Three-node motifs and corresponding themes in the integrated S. cerevisiae network. (a) A motif corresponding to the 'feed-forward' theme; (b) motifs corresponding to the 'co-pointing' theme; (c) motifs corresponding to the 'regulonic complex' theme; (d) motifs corresponding to the 'protein complex' theme; (e) motifs corresponding to the theme of neighborhood clustering of the integrated SSL/homology network; (f) motifs corresponding to the 'compensatory complex members' theme; (g) motifs corresponding to the 'compensatory protein and complex/process' theme; (h) other unclassified motifs. Each of (a-g), from left to right, shows a schematic diagram unifying the collection of motifs in that set, the list of motifs with the motif statistics, a specific example of a subgraph matching one or more of these motifs, and a larger structure corresponding to the network theme. Each colored link represents one of the five interaction types according to the color scheme (bottom right). For a given motif, Nreal is the number of corresponding subgraphs in the real network, and Nrand describes the number of corresponding subgraphs in a randomized network, represented by the average and the standard deviation. A node labeled 'etc.' signifies that the structure contains more nodes with connectivity similar to the labeled node.
Figure 2
Four-node network motifs corresponding to the 'compensatory complexes/processes' theme. (a) A schematic diagram unifying the collection of four-node motifs corresponding to the 'compensatory complexes/processes' theme; (b) examples of specific four-node motifs together with the motif statistics; (c) a specific example of a four-node subgraph matching a few of these motifs; (d) the larger structure corresponding to the network theme. Each colored link represents one of the four interaction types according to the color scheme (see key). For a given motif, Nreal is the number of corresponding subgraphs in the real network, and Nrand describes the number of corresponding subgraphs in a randomized network, represented by the average and the standard deviation.
Figure 3
A thematic map of compensatory complexes. Here, nodes represent protein complexes, and a link is drawn between two nodes if there is a significantly large number of inter-complex SSL interactions. Links between compensatory complexes are labeled with the numbers of supporting SSL interactions.
Figure 4
A thematic map of regulonic complexes. (a) Here, blue nodes represent transcription factors, red nodes represent protein complexes, and a link is drawn between a transcription factor and a protein complex if the promoters of a significantly large number of complex members are bound by the transcription factor. (b) An enlarged region of the regulonic complex map in (a). Links between transcription factors and the complexes they regulate are labeled with the numbers of supporting interactions in the transcription regulation network. For lists of transcription factors and complexes in the map see Additional data files 5 and 6.
Similar articles
- Investigating the Network Basis of Negative Genetic Interactions in Saccharomyces cerevisiae with Integrated Biological Networks and Triplet Motif Analysis.
Ignatius Pang CN, Goel A, Wilkins MR. Ignatius Pang CN, et al. J Proteome Res. 2018 Mar 2;17(3):1014-1030. doi: 10.1021/acs.jproteome.7b00649. Epub 2018 Feb 8. J Proteome Res. 2018. PMID: 29392949 - Untangling the web of functional and physical interactions in yeast.
Herrgård MJ, Palsson BØ. Herrgård MJ, et al. J Biol. 2005;4(2):5. doi: 10.1186/jbiol26. Epub 2005 Jun 8. J Biol. 2005. PMID: 15982410 Free PMC article. Review. - An evolutionary and functional assessment of regulatory network motifs.
Mazurie A, Bottani S, Vergassola M. Mazurie A, et al. Genome Biol. 2005;6(4):R35. doi: 10.1186/gb-2005-6-4-r35. Epub 2005 Mar 24. Genome Biol. 2005. PMID: 15833122 Free PMC article. - Transcriptional regulatory networks in Saccharomyces cerevisiae.
Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK, Young RA. Lee TI, et al. Science. 2002 Oct 25;298(5594):799-804. doi: 10.1126/science.1075090. Science. 2002. PMID: 12399584 - Cell growth and cell cycle in Saccharomyces cerevisiae: basic regulatory design and protein-protein interaction network.
Alberghina L, Mavelli G, Drovandi G, Palumbo P, Pessina S, Tripodi F, Coccetti P, Vanoni M. Alberghina L, et al. Biotechnol Adv. 2012 Jan-Feb;30(1):52-72. doi: 10.1016/j.biotechadv.2011.07.010. Epub 2011 Jul 22. Biotechnol Adv. 2012. PMID: 21821114 Review.
Cited by
- Chemical combination effects predict connectivity in biological systems.
Lehár J, Zimmermann GR, Krueger AS, Molnar RA, Ledell JT, Heilbut AM, Short GF 3rd, Giusti LC, Nolan GP, Magid OA, Lee MS, Borisy AA, Stockwell BR, Keith CT. Lehár J, et al. Mol Syst Biol. 2007;3:80. doi: 10.1038/msb4100116. Epub 2007 Feb 27. Mol Syst Biol. 2007. PMID: 17332758 Free PMC article. - Increasing the power to detect causal associations by combining genotypic and expression data in segregating populations.
Zhu J, Wiener MC, Zhang C, Fridman A, Minch E, Lum PY, Sachs JR, Schadt EE. Zhu J, et al. PLoS Comput Biol. 2007 Apr 13;3(4):e69. doi: 10.1371/journal.pcbi.0030069. Epub 2007 Feb 27. PLoS Comput Biol. 2007. PMID: 17432931 Free PMC article. - Cycle and flow trusses in directed networks.
Takaguchi T, Yoshida Y. Takaguchi T, et al. R Soc Open Sci. 2016 Nov 30;3(11):160270. doi: 10.1098/rsos.160270. eCollection 2016 Nov. R Soc Open Sci. 2016. PMID: 28018610 Free PMC article. - VisANT: an integrative framework for networks in systems biology.
Hu Z, Snitkin ES, DeLisi C. Hu Z, et al. Brief Bioinform. 2008 Jul;9(4):317-25. doi: 10.1093/bib/bbn020. Epub 2008 May 7. Brief Bioinform. 2008. PMID: 18463131 Free PMC article. - Uncovering MicroRNA and Transcription Factor Mediated Regulatory Networks in Glioblastoma.
Sun J, Gong X, Purow B, Zhao Z. Sun J, et al. PLoS Comput Biol. 2012;8(7):e1002488. doi: 10.1371/journal.pcbi.1002488. Epub 2012 Jul 19. PLoS Comput Biol. 2012. PMID: 22829753 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases