Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans - PubMed (original) (raw)
Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans
Gino Poulin et al. EMBO J. 2005.
Erratum in
- EMBO J. 2006 Jan 25;25(2):444-5
Abstract
In Caenorhabditis elegans, numerous 'synMuv' (synthetic multivulval) genes encode for chromatin-associated proteins involved in transcriptional repression, including an orthologue of Rb and components of the NuRD histone deacetylase complex. These genes antagonize Ras signalling to prevent erroneous adoption of vulval fate. To identify new components of this mechanism, we performed a genome-wide RNA interference (RNAi) screen. After RNAi of 16 757 genes, we found nine new synMuv genes. Based on predicted functions and genetic epistasis experiments, we propose that at least four post-translational modifications converge to inhibit Ras-stimulated vulval development: sumoylation, histone tail deacetylation, methylation, and acetylation. In addition, we demonstrate a novel role for sumoylation in inhibiting LIN-12/Notch signalling in the vulva. We further show that many of the synMuv genes are involved in gene regulation outside the vulva, negatively regulating the expression of the Delta homologue lag-2. As most of the genes identified in this screen are conserved in humans, we suggest that similar interactions may be relevant in mammals for control of Ras and Notch signalling, crosstalk between these pathways, and cell proliferation.
Figures
Figure 1
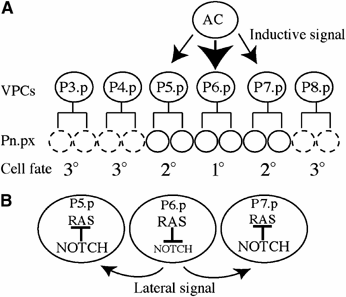
Vulval cell fate specification by the inductive and lateral signals. (A) The inductive signal emanates from the AC. The AC secretes LIN-3 EGF inducing vulval fates in P(5–7).p, which will produce 22 vulval cells. The remaining VPCs have the 3° cell fate, dividing once (dashed circles) and fusing to the hypoderm hyp7. (B) Upon activation of the Ras signalling pathway in P6.p, LIN-12/NOTCH is downregulated in a Ras-dependant manner and its ligands (LAG-2, APX-1, and DSL-1) are expressed, mediating the lateral signal essential for expression of the 2° fate in P5.p and P7.p. In these cells, Notch signalling is activated and negative regulators of Ras are upregulated, reducing Ras signalling.
Figure 2
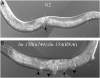
The Muv phenotype. (A) Wild-type N2 adult with normal vulva (arrow). (B) lin-15B(n744); lin-15A(RNAi) adult with multiple vulval protrusions. Arrowheads point to extra vulvae and the arrow to the normal vulva.
Figure 3
HDA-1 is sumoylated. Western blot of wild-type (lane 1) or smo-1 SUMO mutant (lane 2) total worm extract, probed with an anti-HDA-1 antibody. The N2 (WT) extract shows a fast-migrating band at about 65 kDa and a slow-migrating band at about 75 kDa. The slow-migrating band is absent from smo-1 SUMO mutant extract.
Figure 4
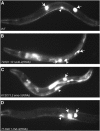
The lag-2_∷_gfp reporter assay. (A) Wild-type mid-L4 stage worm carrying the lag-2_∷_gfp reporter shows strong expression only in the distal tip cells (white arrows) and the vulva (white arrowheads). RNAi of T22D1.10 ruvb-2 helicase (B) or K12C11.2 smo-1 (C) induces strong ectopic expression of lag-2_∷_gfp in the intestine. Only one DTC is visible in the focal plane shown in (C). (D) RNAi of T14G8.1 chd-3 does not induce ectopic lag-2_∷_gfp expression.
Similar articles
- lin-8, which antagonizes Caenorhabditis elegans Ras-mediated vulval induction, encodes a novel nuclear protein that interacts with the LIN-35 Rb protein.
Davison EM, Harrison MM, Walhout AJ, Vidal M, Horvitz HR. Davison EM, et al. Genetics. 2005 Nov;171(3):1017-31. doi: 10.1534/genetics.104.034173. Epub 2005 Jul 14. Genetics. 2005. PMID: 16020796 Free PMC article. - Sumoylation of LIN-1 promotes transcriptional repression and inhibition of vulval cell fates.
Leight ER, Glossip D, Kornfeld K. Leight ER, et al. Development. 2005 Mar;132(5):1047-56. doi: 10.1242/dev.01664. Epub 2005 Feb 2. Development. 2005. PMID: 15689373 - dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signaling in C. elegans vulval development.
Ceol CJ, Horvitz HR. Ceol CJ, et al. Mol Cell. 2001 Mar;7(3):461-73. doi: 10.1016/s1097-2765(01)00194-0. Mol Cell. 2001. PMID: 11463372 - Signal transduction and cell fate specification during Caenorhabditis elegans vulval development.
Eisenmann DM, Kim SK. Eisenmann DM, et al. Curr Opin Genet Dev. 1994 Aug;4(4):508-16. doi: 10.1016/0959-437x(94)90065-b. Curr Opin Genet Dev. 1994. PMID: 7950317 Review. - Pattern formation during C. elegans vulval induction.
Wang M, Sternberg PW. Wang M, et al. Curr Top Dev Biol. 2001;51:189-220. doi: 10.1016/s0070-2153(01)51006-6. Curr Top Dev Biol. 2001. PMID: 11236714 Review.
Cited by
- Combinatorial RNA interference in Caenorhabditis elegans reveals that redundancy between gene duplicates can be maintained for more than 80 million years of evolution.
Tischler J, Lehner B, Chen N, Fraser AG. Tischler J, et al. Genome Biol. 2006;7(8):R69. doi: 10.1186/gb-2006-7-8-R69. Epub 2006 Aug 2. Genome Biol. 2006. PMID: 16884526 Free PMC article. - Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation.
Nie M, Xie Y, Loo JA, Courey AJ. Nie M, et al. PLoS One. 2009 Jun 16;4(6):e5905. doi: 10.1371/journal.pone.0005905. PLoS One. 2009. PMID: 19529778 Free PMC article. - Biology of the Mi-2/NuRD Complex in SLAC (Stemness, Longevity/Ageing, and Cancer).
Zhang Y. Zhang Y. Gene Regul Syst Bio. 2011 Feb 27;5:1-26. doi: 10.4137/GRSB.S6510. Gene Regul Syst Bio. 2011. PMID: 21523247 Free PMC article. - Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt).
Tian Y, Garcia G, Bian Q, Steffen KK, Joe L, Wolff S, Meyer BJ, Dillin A. Tian Y, et al. Cell. 2016 May 19;165(5):1197-1208. doi: 10.1016/j.cell.2016.04.011. Epub 2016 Apr 28. Cell. 2016. PMID: 27133166 Free PMC article. - Keep quiet: the HUSH complex in transcriptional silencing and disease.
Müller I, Helin K. Müller I, et al. Nat Struct Mol Biol. 2024 Jan;31(1):11-22. doi: 10.1038/s41594-023-01173-7. Epub 2024 Jan 12. Nat Struct Mol Biol. 2024. PMID: 38216658 Review.
References
- Aroian RV, Koga M, Mendel JE, Ohshima Y, Sternberg PW (1990) The let-23 gene necessary for Caenorhabditis elegans vulval induction encodes a tyrosine kinase of the EGF receptor subfamily. Nature 348: 693–699 - PubMed
- Beitel GJ, Clark SG, Horvitz HR (1990) Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature 348: 503–509 - PubMed
- Beitel GJ, Tuck S, Greenwald I, Horvitz HR (1995) The Caenorhabditis elegans gene lin-1 encodes an ETS-domain protein and defines a branch of the vulval induction pathway. Genes Dev 9: 3149–3162 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials