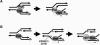Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands - PubMed (original) (raw)
Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands
Colin P Guy et al. Nucleic Acids Res. 2005.
Abstract
Mutations in mammalian and Drosophila Hel308 and PolQ paralogues cause genome instability but their helicase functions are mysterious. By in vivo and in vitro analysis, we show that Hel308 from archaea (Hel308a) may act at stalled replication forks. Introducing hel308a into Escherichia coli dnaE strains that conditionally accumulate stalled forks caused synthetic lethality, an effect indistinguishable from E.coli RecQ. Further analysis in vivo indicated that the effect of hel308a is exerted independently of homologous recombination. The minimal biochemical properties of Hel308a protein were the same as human Hel308. We describe how helicase actions of Hel308a at fork structures lead specifically to displacement of lagging strands. The invading strand of D-loops is also targeted. Using archaeal Hel308, we propose models of action for the helicase domain of PolQ, promoting loading of the translesion polymerase domain. We speculate that removal of lagging strands at stalled forks by Hel308 promotes the formation of initiation zones, priming restart of lagging strand synthesis.
Figures
Figure 1
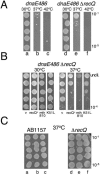
Hel308 from archaea (mth810) interacts genetically with stalled replication forks. (A) Serial dilution of overnight cultures grown at 30°C showing growth of E.coli dnaE486 (lane a) that is severely restricted at 37°C (lane b) is improved by Δ_recQ_ (lane e). (B) Spot tests from overnight cultures grown at 30°C, showing that plasmid encoded archaeal gene mth810 stunts growth of dnaE486 Δ_recQ_ at 37°C, the same phenotype as plasmid encoded E.coli recQ. V is empty vector control and K51L denotes the mutation introduced into helicase motif I of mth810 (C) Mth810 has no dominant effect when transformed into wild-type or Δ_recQ_ strains; lanes a and d are empty vector; lanes b and e are recQ+; and lanes c and f are mth810.
Figure 2
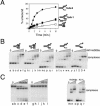
Mth810 is the archaeal orthologue of metazoan Hel308 in sequence and minimal helicase function. (A) Cartoon showing common features of Hel308 from archaea (Hel308a), human (hHel308) and the N-terminal domain of human PolQ. Helicase motifs, including the Q-motif (53), are labelled and the Hel308a sequences are given for motif I and IVa with mutagenized residues in bold and underlined. (B) Sequence details in helicase motifs V and VI that confirm Hel308a as a Hel308/Mus308 family rather than a RecQ helicase. The corresponding motif of human BLM helicase is shown for comparison (hBLM). In each motif peculiar residues conserved in Hel308/Mus308 helicases are in bold. Motif IVa is highly conserved in RecQ and Hel308 proteins. Invariant residues are in bold and highly conserved residues are underlined. Aligned with Hel308a, human Hel308 and human PolQ are Hel308 from Caenorhabditis elegans (CeHel308), E.coli RecQ (EcRecQ) and a human RecQ, BLM (HsBLM). (C) SDS–PAGE gel (10% acrylamide) showing purified recombinant Hel308a (arrowed) from Methanothermobacter. Marker sizes are given on the left of the panel. (D) ATPase activity of Hel308a measured as a function of time in reactions containing no DNA (filled diamond), dsDNA (open square) or ssDNA (open circles). Error bars are derived from the means of three independent experiments. (E) Unwinding reactions of Hel308a on 3′-ssDNA-tailed duplex (i), 5′-ssDNA-tailed duplex (ii) and untailed duplex (iii). Reactions were for 20 min at 45°C containing 2 nM DNA, with 32P-labelled strand indicated by filled circle, 5 mM MgCl2, 5 mM ATP and zero (lane a); 1, 5, 10, 25 and 50 nM Hel308a (lanes b–f).
Figure 3
Hel308 from archaea preferentially targets fork DNA for unwinding. (A) Gel-retardation binding assays of Hel308a on the substrates in Figure 2E in 1 mM magnesium. Reactions were at 45°C for 10 min and contained 2 nM DNA substrate mixed with 0, 1, 2, 10, 50 and 100 nM Hel308a. (B) The same reactions as in (A), but containing an additional 1 mM ATP in the gel, and all buffers. (C). Unwinding reactions of Hel308a on flayed duplex (lanes a–f), fork with leading strand only (lanes g–l), fork with lagging strand only (fork-2, lanes m–r), fork with both leading and lagging strands (fork-1, lanes s–x) and Holliday junction (lanes y–dd). Reactions were for 20 min at 45°C containing 2 nM DNA, 5 mM MgCl2, 5 mM ATP and zero (lanes a, g, m, s and y) or 1, 5, 10, 25 and 50 nM Hel308a.
Figure 4
Hel308a targets DNA forks for unwinding and binds to duplex substrates with branchpoints. (A) Time-course unwinding of four substrates acted on by Hel308a: fork-2 (filled circles), fork-1 (filled squares), 3′-ssDNA-tailed duplex (open circles) and Holliday junction (open squares). Substrates used are annotated to the right of the graph. Reactions were at 45°C for the times shown and contained 2 nM DNA, 5 mM MgCl2, 5 mM ATP and 20 nM Hel308a. Error bars are mostly hidden by data points and were derived from means of three independent assays. (B) Gel-retardation binding assays of Hel308a on fully base-paired, static Holliday junction (lanes a–f), static Holliday junction containing a backbone nick indicated by an arrowhead (lanes g–l), fully base-paired fork-3 (lanes m–r), fork-3 lacking a leading strand (lanes s–x) and fork-3 lacking a lagging strand (lanes y–4). Reactions were at 45°C for 10 min in 1 mM magnesium and contained 2 nM DNA and Hel308a at 0, 1, 2, 10, 50 and100 nM. (C) Hel308a K51L protein is unable to unwind fork-2 (lanes g–l), compared with wild-type protein (lanes a–f). Reactions were for 20 min at 45°C containing 2 nM DNA, 5 mM MgCl2, 5 mM ATP and 0, 1, 5, 10, 25 or 50 nM Hel308a. Binding of wild-type Hel308a (lanes m–o) and K51L Hel308a (lanes p–r) to fork 2 were in reactions for 10 min at 45°C containing 5 mM MgCl2, 2 nM fork-2 (labelled on strand 1) and 0, 10 or 100 nM Hel308a protein.
Figure 5
Hel308 from archaea (mth810) targets different substrates from RuvABC in vivo. (A) Typical spot test plate of the growth phenotype from expression of mth810 in E.coli Δ_ruvABC_ exposed to 0 or 20 J/m2 UV light. (B and C) Effect mth810 (pEB310) on UV survival of wild-type E.coli (MG1655, circles), Δ_recG_ (squares) and Δ_ruvABC_ (triangles) strains (B), and a Δ_recG_ Δ_ruvABC_ strain (C). Graphs represent means of three independent experiments.
Figure 6
Hel308a unwinds lagging strands from nicks and the fork branchpoint. (A) Products from unwinding fork-2 by hel308a as a function of time. Reactions contained 2 nM DNA and 20 nM Hel308a in 5 mM MgCl2, 5 mM ATP at 45°C. (B) Reactions showing unwinding of strands in fork-1 by Hel308a. Cartoons of the substrate are shown above the panel in each case with the labelled strand denoted by a filled circle. Strands are numbered on one of these cartoons. Reactions were at 45°C for 20 min containing 5 mM MgCl2, 5 mM ATP, 2 nM DNA and zero (0) or 50 nM Hel308a (H). Lanes marked B contained no Hel308a and the reactions were heated to 95°C for 20 min. Letters X, Y and Z highlight the major product of unwinding in each reaction containing Hel308a. Substrate markers corresponding to fork lacking a lagging strand (M1), flayed duplex (M2) and partial duplex (M3) are annotated beside the panel. (C) Time-course unwinding of fork-3 and its corresponding nicked duplex DNA substrate. Substrates used are annotated to the right of the graph. Reactions were at 45°C for the times shown and contained 2 nM DNA, 5 mM MgCl2, 5 mM ATP and 20 nM Hel308a. Error bars derive from means of two independent assays. (D) Unwinding of the invading strand of D-loop substrates by Hel308a. The D-loop substrates used are annotated above the panel and reactions products are displayed to the right of the panel. Reactions were for 20 min at 45°C containing 2 nM DNA, 5 mM MgCl2, 5 mM ATP and zero or 1, 5, 10, 25 and 50 nM Hel308a.
Figure 7
Models proposing how Hel308 helicases may function in archaea and metazoans. (A) Displacement of the lagging strand by the Hel308 helicase (Hel) domain of PolQ may provide access for the translesion polymerase domain to DNA damage (grey square, e.g. AP-sites) located on the lagging strand template. (B) Hel308a/Hel308 (Hel) unwinding of the lagging strand at fork with a compromised leading or lagging strand provides a template for loading of replication restart apparatus, possibly Pol-α-primase (αPri). In each model, translocation of the helicase is indicted by a dotted arrow, away from the fork branchpoint in a 3′–5′ direction with respect to the lagging strand template.
Similar articles
- Human HEL308 localizes to damaged replication forks and unwinds lagging strand structures.
Tafel AA, Wu L, McHugh PJ. Tafel AA, et al. J Biol Chem. 2011 May 6;286(18):15832-40. doi: 10.1074/jbc.M111.228189. Epub 2011 Mar 11. J Biol Chem. 2011. PMID: 21398521 Free PMC article. - DNA binding and unwinding by Hel308 helicase requires dual functions of a winged helix domain.
Northall SJ, Buckley R, Jones N, Penedo JC, Soultanas P, Bolt EL. Northall SJ, et al. DNA Repair (Amst). 2017 Sep;57:125-132. doi: 10.1016/j.dnarep.2017.07.005. Epub 2017 Jul 16. DNA Repair (Amst). 2017. PMID: 28738244 - Hjm/Hel308A DNA helicase from Sulfolobus tokodaii promotes replication fork regression and interacts with Hjc endonuclease in vitro.
Li Z, Lu S, Hou G, Ma X, Sheng D, Ni J, Shen Y. Li Z, et al. J Bacteriol. 2008 Apr;190(8):3006-17. doi: 10.1128/JB.01662-07. Epub 2008 Feb 22. J Bacteriol. 2008. PMID: 18296528 Free PMC article. - Molecular biology of Hel308 helicase in archaea.
Woodman IL, Bolt EL. Woodman IL, et al. Biochem Soc Trans. 2009 Feb;37(Pt 1):74-8. doi: 10.1042/BST0370074. Biochem Soc Trans. 2009. PMID: 19143605 Review. - Helicases that interact with replication forks: new candidates from archaea.
Bolt EL. Bolt EL. Biochem Soc Trans. 2005 Dec;33(Pt 6):1471-3. doi: 10.1042/BST0331471. Biochem Soc Trans. 2005. PMID: 16246148 Review.
Cited by
- Structure of the Helicase Domain of DNA Polymerase Theta Reveals a Possible Role in the Microhomology-Mediated End-Joining Pathway.
Newman JA, Cooper CDO, Aitkenhead H, Gileadi O. Newman JA, et al. Structure. 2015 Dec 1;23(12):2319-2330. doi: 10.1016/j.str.2015.10.014. Structure. 2015. PMID: 26636256 Free PMC article. - Remodeling and Control of Homologous Recombination by DNA Helicases and Translocases that Target Recombinases and Synapsis.
Northall SJ, Ivančić-Baće I, Soultanas P, Bolt EL. Northall SJ, et al. Genes (Basel). 2016 Aug 19;7(8):52. doi: 10.3390/genes7080052. Genes (Basel). 2016. PMID: 27548227 Free PMC article. Review. - Revealing dynamics of helicase translocation on single-stranded DNA using high-resolution nanopore tweezers.
Craig JM, Laszlo AH, Brinkerhoff H, Derrington IM, Noakes MT, Nova IC, Tickman BI, Doering K, de Leeuw NF, Gundlach JH. Craig JM, et al. Proc Natl Acad Sci U S A. 2017 Nov 7;114(45):11932-11937. doi: 10.1073/pnas.1711282114. Epub 2017 Oct 16. Proc Natl Acad Sci U S A. 2017. PMID: 29078357 Free PMC article. - Structure and Function of a Novel ATPase that Interacts with Holliday Junction Resolvase Hjc and Promotes Branch Migration.
Zhai B, DuPrez K, Doukov TI, Li H, Huang M, Shang G, Ni J, Gu L, Shen Y, Fan L. Zhai B, et al. J Mol Biol. 2017 Apr 7;429(7):1009-1029. doi: 10.1016/j.jmb.2017.02.016. Epub 2017 Feb 24. J Mol Biol. 2017. PMID: 28238763 Free PMC article. - The conjugative DNA translocase TrwB is a structure-specific DNA-binding protein.
Matilla I, Alfonso C, Rivas G, Bolt EL, de la Cruz F, Cabezon E. Matilla I, et al. J Biol Chem. 2010 Jun 4;285(23):17537-44. doi: 10.1074/jbc.M109.084137. Epub 2010 Apr 6. J Biol Chem. 2010. PMID: 20375020 Free PMC article.
References
- Aylon Y., Kupiec M. DSB repair: the yeast paradigm. DNA Repair (Amst.) 2004;3:797–815. - PubMed
- Cox M.M., Goodman M.F., Kreuzer K.N., Sherratt D.J., Sandler S.J., Marians K.J. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. - PubMed
- Michel B. Replication fork arrest and DNA recombination. Trends Biochem. Sci. 2000;25:173–178. - PubMed
- McGlynn P., Lloyd R.G. Recombinational repair and restart of damaged replication forks. Nature Rev. Mol. Cell Biol. 2002;3:859–870. - PubMed