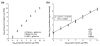Transcript copy number estimation using a mouse whole-genome oligonucleotide microarray - PubMed (original) (raw)
Transcript copy number estimation using a mouse whole-genome oligonucleotide microarray
Mark G Carter et al. Genome Biol. 2005.
Abstract
The ability to quantitatively measure the expression of all genes in a given tissue or cell with a single assay is an exciting promise of gene-expression profiling technology. An in situ-synthesized 60-mer oligonucleotide microarray designed to detect transcripts from all mouse genes was validated, as well as a set of exogenous RNA controls derived from the yeast genome (made freely available without restriction), which allow quantitative estimation of absolute endogenous transcript abundance.
Figures
Figure 1
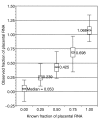
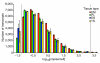
60-mer oligonucleotide probe linearity testing. To test the performance of 21,044 60-mer oligonucleotide probes, E12.5 embryo RNA and placenta RNA were combined to form five pairs of duplicate samples containing from 0 to 100% placental RNA. Box-plot distribution data for each placental RNA input level is shown above, with median values labeled. The boxes show the 25-75 percentile range, with the mean and median indicated by the central straight line and diamond, respectively. Upper and lower bars show the 2.5 to 97.5 percentile range. Observed fraction medians are within 0.075 of input values, and 95% of values are within 0.405 of input values.
Figure 2
Relating yeast spike-in RNA control copy number to qPCR measurements and microarray signal intensity. (a) To verify abundances of yeast sequence RNA transcripts in a control mixture, cDNA was transcribed from the control mixture alone (open boxes), as well as E12.5 whole-mouse embryo total RNA (open diamonds) and Universal Mouse RNA (filled triangles) with added spike-in control mixture. The cDNA was used as template for real-time PCR quantitation of each yeast sequence RNA, using a separately prepared standard of cDNA transcribed from the yeast sequences. Expected and measured copy numbers are closely matched (_r_2 ≥ 0.99), with maximum measured/observed ratios of 1.5, 1.5, and 2.6, respectively. (b) Expression profiles were generated for triplicate total RNA samples from E12.5 embryo (filled circles), E12.5 placenta (open circles), ES cells (filled boxes), and TS cells (open boxes) with yeast sequence control transcripts spiked-in prior to target labeling. For the seven control transcripts, mean log10[intensity] is shown for each tissue type, as well as the mean across all samples (filled triangles), and these data were used to perform linear regression analysis and relate signal intensity to transcript copy number, allowing abundance estimation for endogenous transcripts. The regression line for the average of all tissues (dashed line) and its equation is shown. Intensity-copy number correlations for individual tissues were very strong, with _r_2 values of 0.98 - 0.99.
Figure 3
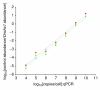
Exogenous control and endogenous transcript amplification rates are closely matched over seven orders of magnitude. Transcript abundance of each spike-in control transcript was measured by qPCR before and after linear amplification labeling, and compared to amounts of the exogenous transcript Dnchc1. After amplification, individual ratios of each control transcript to the endogenous transcript were within 3.5-fold (average = 1.98-fold) of those prior to amplification. Blue diamonds = log10[ratio mean control/Dnchc1 transcripts] of three E12.5 embryo and three E12.5 placenta samples before amplification. Red boxes, green triangles = log10[ratio mean control/Dnchc1 transcripts] for the same samples after amplification, using yield versus input (red boxes) or the increase in Dnchc1 transcripts as measured by qPCR (green triangles) to calculate the fraction of the original sample represented by each qPCR well.
Figure 4
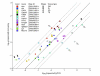
Validation of transcript abundance estimation for endogenous transcripts. qPCR primer sets were designed for selected genes so that amplicons were upstream of 60-mer oligonucleotide probes when possible, or less than 650 bp downstream, and copy number was estimated using serial dilutions of RNA, in vitro transcribed from mouse cDNAs, at known copy numbers as standards. Error bars represent one standard deviation across three replicate samples for each tissue. Dotted diagonal lines represent five- and tenfold differences between the two datasets. Each gene's official symbol, along with the unique identifier for the 60-mer oligonucleotide probe it was measured with, are listed in the key. Data was normalized to Gapd expression for both methods. EM = E12.5 embryo, PL = E12.5 placenta, ES = embryonic stem cells, TS = trophoblast stem cells.
Figure 5
Distribution of mouse transcript abundances in E12.5 embryo and placenta, and cultured ES and TS cells. Transcript abundances are expressed as log10[copies per cell], varying over six orders of magnitude. The distributions are highly similar, despite the significant differences between the four tissues (for example, monolayer culture versus tissue, placenta versus embryo), suggesting that such distributions are not heavily skewed according to tissue structure or function. The percentage of transcripts present at less than one copy per cell ranged from 40.1 to 48.2% in the four tissues. Bins were centered on indicated values, and the dotted lines indicate values corresponding to mean upper and lower signal intensity reliability limits of one copy per 26 cells to 2,188 copies per cell. For definitions of tissue type see Figure 4 legend.
Similar articles
- Expression profiling of the bottom fermenting yeast Saccharomyces pastorianus orthologous genes using oligonucleotide microarrays.
Minato T, Yoshida S, Ishiguro T, Shimada E, Mizutani S, Kobayashi O, Yoshimoto H. Minato T, et al. Yeast. 2009 Mar;26(3):147-65. doi: 10.1002/yea.1654. Yeast. 2009. PMID: 19243081 - A generic approach for the design of whole-genome oligoarrays, validated for genomotyping, deletion mapping and gene expression analysis on Staphylococcus aureus.
Charbonnier Y, Gettler B, François P, Bento M, Renzoni A, Vaudaux P, Schlegel W, Schrenzel J. Charbonnier Y, et al. BMC Genomics. 2005 Jun 17;6:95. doi: 10.1186/1471-2164-6-95. BMC Genomics. 2005. PMID: 15963225 Free PMC article. - A novel design of whole-genome microarray probes for Saccharomyces cerevisiae which minimizes cross-hybridization.
Talla E, Tekaia F, Brino L, Dujon B. Talla E, et al. BMC Genomics. 2003 Sep 22;4(1):38. doi: 10.1186/1471-2164-4-38. BMC Genomics. 2003. PMID: 14499002 Free PMC article. - Global analysis of gene expression in yeast.
Horak CE, Snyder M. Horak CE, et al. Funct Integr Genomics. 2002 Sep;2(4-5):171-80. doi: 10.1007/s10142-002-0065-3. Epub 2002 Jul 10. Funct Integr Genomics. 2002. PMID: 12192590 Review. - [Transcriptomes for serial analysis of gene expression].
Marti J, Piquemal D, Manchon L, Commes T. Marti J, et al. J Soc Biol. 2002;196(4):303-7. J Soc Biol. 2002. PMID: 12645300 Review. French.
Cited by
- Prenatal arsenic exposure alters gene expression in the adult liver to a proinflammatory state contributing to accelerated atherosclerosis.
States JC, Singh AV, Knudsen TB, Rouchka EC, Ngalame NO, Arteel GE, Piao Y, Ko MS. States JC, et al. PLoS One. 2012;7(6):e38713. doi: 10.1371/journal.pone.0038713. Epub 2012 Jun 15. PLoS One. 2012. PMID: 22719926 Free PMC article. - Pan-genome isolation of low abundance transcripts using SAGE tag.
Kim YC, Jung YC, Xuan Z, Dong H, Zhang MQ, Wang SM. Kim YC, et al. FEBS Lett. 2006 Dec 11;580(28-29):6721-9. doi: 10.1016/j.febslet.2006.11.013. Epub 2006 Nov 14. FEBS Lett. 2006. PMID: 17113583 Free PMC article. - Effects of maternal nutrient restriction during the periconceptional period on placental development in the mouse.
Van Gronigen Case G, Storey KM, Parmeley LE, Schulz LC. Van Gronigen Case G, et al. PLoS One. 2021 Jan 14;16(1):e0244971. doi: 10.1371/journal.pone.0244971. eCollection 2021. PLoS One. 2021. PMID: 33444393 Free PMC article. - Maintenance of undifferentiated mouse embryonic stem cells in suspension by the serum- and feeder-free defined culture condition.
Tsuji Y, Yoshimura N, Aoki H, Sharov AA, Ko MS, Motohashi T, Kunisada T. Tsuji Y, et al. Dev Dyn. 2008 Aug;237(8):2129-38. doi: 10.1002/dvdy.21617. Dev Dyn. 2008. PMID: 18624284 Free PMC article. - An in situ hybridization-based screen for heterogeneously expressed genes in mouse ES cells.
Carter MG, Stagg CA, Falco G, Yoshikawa T, Bassey UC, Aiba K, Sharova LV, Shaik N, Ko MS. Carter MG, et al. Gene Expr Patterns. 2008 Feb;8(3):181-98. doi: 10.1016/j.gep.2007.10.009. Epub 2007 Nov 4. Gene Expr Patterns. 2008. PMID: 18178135 Free PMC article.
References
- Schadt EE, Edwards SW, GuhaThakurta D, Holder D, Ying L, Svetnik V, Leonardson A, Hart KW, Russell A, Li G, et al. A comprehensive transcript index of the human genome generated using microarrays and computational approaches. Genome Biol. 2004;5:R73. doi: 10.1186/gb-2004-5-10-r73. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous