Reevaluation of the role of the med-1 and med-2 genes in specifying the Caenorhabditis elegans endoderm - PubMed (original) (raw)
Comparative Study
Reevaluation of the role of the med-1 and med-2 genes in specifying the Caenorhabditis elegans endoderm
Barbara Goszczynski et al. Genetics. 2005 Oct.
Abstract
The med-1 and med-2 genes encode a pair of essentially identical GATA factor-related transcription factors that have been proposed to be necessary for specification of the C. elegans endoderm (intestine or E lineage) as well as part of the C. elegans mesoderm. med-1 and med-2 are proposed to be the direct downstream targets and the principal effectors of the maternally provided SKN-1 transcription factor; med-1 and med-2 would thus occupy the pivotal interface between maternal and zygotic control of gene expression. The conclusion that med-1 and med-2 are necessary for C. elegans endoderm specification was based on a partially penetrant (approximately 50%) loss of endoderm markers produced by RNA-mediated interference (RNAi). To determine whether this partial penetrance reflects: (i) inefficient RNAi against early zygotic transcripts, (ii) experimental uncertainty in the expected level of endoderm loss in skn-1 nulls, or (iii) additional redundancy in the pathway of endoderm specification, we constructed worm strains that segregate embryos lacking both the med-1 gene (because of a gene-specific deletion) and the med-2 gene (using either of two chromosomal deficiencies). Contrary to expectations, we observe that only approximately 3-20% of med-2(-); med-1(-) embryos do not express markers of endoderm differentiation. Furthermore, we found no evidence for a maternal contribution of the med genes to endoderm specification. We conclude that the major pathway(s) for endoderm specification in C. elegans must be independent of the med-1 and med-2 genes.
Figures
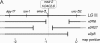
Figure 1.
Cell lineage of the early C. elegans embryo (left), aligned with the proposed transcription factor cascade that leads to specification of the C. elegans endoderm (right). Lineages that lead to the intestine are solid; other lineages are shaded. Only transcription factors that are on the proposed endoderm specification pathway are shown; in particular, roles for SKN-1 and MED-1,2 in specification of the MS lineage are not shown. The proposed activation by SKN-1 of the med-1 and med-2 genes marks the transition from maternal to zygotic control of gene expression. This figure was redrawn from Figure 4 of M
aduro
and R
othman
(2002).
Figure 2.
Genetic positions and molecular characteristics of the med-1 and med-2 genes. (A) The genetic map of the middle of the C. elegans chromosome III. The med-2 gene is located on cosmid K04C2, which is removed by the chromosomal deficiencies sDf127 and nDf16. As described in the text, the free duplication sDp3 is used to balance the deficiency sDf127; a chromosome marked with lon-1 unc-32 is used to balance the deficiency nDf16. (B) Expanded views of the chromosomal regions surrounding the med-2 gene (top) and the med-1 gene (bottom). Coordinates correspond to the cosmid sequences K04C2 and T24D3 for med-2 and med-1, respectively. The intronless ORFs corresponding to med-1 and med-2 are indicated by the solid boxes. The shaded bars above the med-2 and med-1 ORFs indicate the chromosomal regions that show high sequence similarity between the two genes (see M
aduro
et al. 2001 for details). Because of this region of high sequence similarity, the presence of med-1 and med-2 genes can be assayed simultaneously and independently by the same set of primers, oJM296 and oJM297. The extent of the med-1 deletion allele ok804 is indicated. We note that an alternative structure of the med-1 gene (containing a 3′-intron) has previously appeared in the C. elegans sequence annotation: the ok804 allele removes the predicted med-1 zinc-finger DNA-binding domain and would be predicted to produce a genetic null for either gene model. The ok804 deletion is associated with a 790-bp insertion of a (non-ORF-containing) sequence from cosmid C44C10, allowing the deletion to be detected by PCR primers oJM304 and oJM305, as indicated. The cross-hatched region beneath the med-2 gene was amplified by PCR, cloned, and used as a probe to detect both med-2 and med-1 sequences by Southern blotting on genomic DNA digested with the restriction enzyme StyI. (C) PCR detection of med-1(+), med-1(ok804), and med-2(+) alleles in individual arrested embryos produced by strains N2 (wild type), RB930 [med-1(ok804)], JM134 med-1(−), and JM133 med-1(+). The deduced status of the med genes in the individual embryos are indicated beside the strain names. Locations of PCR primers within the med-1 and med-2 genes are shown in B.
Figure 2.
Genetic positions and molecular characteristics of the med-1 and med-2 genes. (A) The genetic map of the middle of the C. elegans chromosome III. The med-2 gene is located on cosmid K04C2, which is removed by the chromosomal deficiencies sDf127 and nDf16. As described in the text, the free duplication sDp3 is used to balance the deficiency sDf127; a chromosome marked with lon-1 unc-32 is used to balance the deficiency nDf16. (B) Expanded views of the chromosomal regions surrounding the med-2 gene (top) and the med-1 gene (bottom). Coordinates correspond to the cosmid sequences K04C2 and T24D3 for med-2 and med-1, respectively. The intronless ORFs corresponding to med-1 and med-2 are indicated by the solid boxes. The shaded bars above the med-2 and med-1 ORFs indicate the chromosomal regions that show high sequence similarity between the two genes (see M
aduro
et al. 2001 for details). Because of this region of high sequence similarity, the presence of med-1 and med-2 genes can be assayed simultaneously and independently by the same set of primers, oJM296 and oJM297. The extent of the med-1 deletion allele ok804 is indicated. We note that an alternative structure of the med-1 gene (containing a 3′-intron) has previously appeared in the C. elegans sequence annotation: the ok804 allele removes the predicted med-1 zinc-finger DNA-binding domain and would be predicted to produce a genetic null for either gene model. The ok804 deletion is associated with a 790-bp insertion of a (non-ORF-containing) sequence from cosmid C44C10, allowing the deletion to be detected by PCR primers oJM304 and oJM305, as indicated. The cross-hatched region beneath the med-2 gene was amplified by PCR, cloned, and used as a probe to detect both med-2 and med-1 sequences by Southern blotting on genomic DNA digested with the restriction enzyme StyI. (C) PCR detection of med-1(+), med-1(ok804), and med-2(+) alleles in individual arrested embryos produced by strains N2 (wild type), RB930 [med-1(ok804)], JM134 med-1(−), and JM133 med-1(+). The deduced status of the med genes in the individual embryos are indicated beside the strain names. Locations of PCR primers within the med-1 and med-2 genes are shown in B.
Figure 2.
Genetic positions and molecular characteristics of the med-1 and med-2 genes. (A) The genetic map of the middle of the C. elegans chromosome III. The med-2 gene is located on cosmid K04C2, which is removed by the chromosomal deficiencies sDf127 and nDf16. As described in the text, the free duplication sDp3 is used to balance the deficiency sDf127; a chromosome marked with lon-1 unc-32 is used to balance the deficiency nDf16. (B) Expanded views of the chromosomal regions surrounding the med-2 gene (top) and the med-1 gene (bottom). Coordinates correspond to the cosmid sequences K04C2 and T24D3 for med-2 and med-1, respectively. The intronless ORFs corresponding to med-1 and med-2 are indicated by the solid boxes. The shaded bars above the med-2 and med-1 ORFs indicate the chromosomal regions that show high sequence similarity between the two genes (see M
aduro
et al. 2001 for details). Because of this region of high sequence similarity, the presence of med-1 and med-2 genes can be assayed simultaneously and independently by the same set of primers, oJM296 and oJM297. The extent of the med-1 deletion allele ok804 is indicated. We note that an alternative structure of the med-1 gene (containing a 3′-intron) has previously appeared in the C. elegans sequence annotation: the ok804 allele removes the predicted med-1 zinc-finger DNA-binding domain and would be predicted to produce a genetic null for either gene model. The ok804 deletion is associated with a 790-bp insertion of a (non-ORF-containing) sequence from cosmid C44C10, allowing the deletion to be detected by PCR primers oJM304 and oJM305, as indicated. The cross-hatched region beneath the med-2 gene was amplified by PCR, cloned, and used as a probe to detect both med-2 and med-1 sequences by Southern blotting on genomic DNA digested with the restriction enzyme StyI. (C) PCR detection of med-1(+), med-1(ok804), and med-2(+) alleles in individual arrested embryos produced by strains N2 (wild type), RB930 [med-1(ok804)], JM134 med-1(−), and JM133 med-1(+). The deduced status of the med genes in the individual embryos are indicated beside the strain names. Locations of PCR primers within the med-1 and med-2 genes are shown in B.
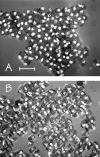
Figure 3.
Microscopic images of typical arrested embryos produced by control strain JM133 med-1(+) (A and B) and by strain JM134 med-1(−) (C and D), as seen by differential interference contrast (A and C) or by birefringence optics (B and D). As described in the text, the majority of the arrested embryos produced by either strain still express birefringent gut granules. The arrows in C and D indicate one arrested embryo produced by strain JM134 med-1(−) that does not express gut granules. Bar, 50 μm. This experiment was repeated six times for each genotype; the total number of scored embryos was 671 and 690 for strains JM133 med-1(+) and JM134 med-1(−), respectively.
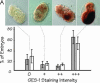
Figure 4.
Expression of additional markers of endoderm differentiation in arrested embryos produced by strains JM133 med-1(+) and JM134 med-1(−). (A) Histogram showing the distribution of GES-1 staining intensity (classified as 0, +, ++, or +++) within arrested embryos produced by strain JM134 med-1(−) (open bars) or by control strain JM133 med-1(+) (shaded bars). Typical images of each class of stained embryo [from strain JM134 med-1(−)] are shown above the corresponding histogram bar. The experiment was repeated a total of five times for each genotype, scoring a total of 830 and 872 individual arrested embryos produced by strains JM133 med-1(+) and JM134 med-1(−), respectively; error bars are standard deviations. The apparent difference between the JM133 med-1(+) and JM134 med-1(−) histograms is due entirely to one particular pair of slides (corresponding to the longest incubation time) in which some embryos appeared to have degenerated and the staining appeared erratic. If this set of data is excluded from the analysis, the behaviors of JM133 med-1(+) and JM134 med-1(−) are essentially indistinguishable (data not shown). (B) Histogram showing the distribution of MH33 staining intensity (classified as 0, +, ++) within arrested embryos produced by strain JM134 med-1(−) (open bars) or control strain JM133 med-1(+) (shaded bars). Images [from strain JM134 med-1(−)] represent Z-projections of deconvolved image stacks taken for each embryo; blue, DAPI staining; red, MH33 immunofluorescence. A well-formed endotube is obvious in embryos classified as “++”; a rudimentary endotube can be detected in embryos classified as “+” (see arrow). This experiment was repeated five times, scoring a total of 305 and 386 arrested embryos for strains JM133 med-1(+) and JM134 med-1(−), respectively; error bars represent standard deviations. DAPI-stained images from these experiments were used to estimate the total number of nuclei in the arrested embryos (Table 1).
Figure 4.
Expression of additional markers of endoderm differentiation in arrested embryos produced by strains JM133 med-1(+) and JM134 med-1(−). (A) Histogram showing the distribution of GES-1 staining intensity (classified as 0, +, ++, or +++) within arrested embryos produced by strain JM134 med-1(−) (open bars) or by control strain JM133 med-1(+) (shaded bars). Typical images of each class of stained embryo [from strain JM134 med-1(−)] are shown above the corresponding histogram bar. The experiment was repeated a total of five times for each genotype, scoring a total of 830 and 872 individual arrested embryos produced by strains JM133 med-1(+) and JM134 med-1(−), respectively; error bars are standard deviations. The apparent difference between the JM133 med-1(+) and JM134 med-1(−) histograms is due entirely to one particular pair of slides (corresponding to the longest incubation time) in which some embryos appeared to have degenerated and the staining appeared erratic. If this set of data is excluded from the analysis, the behaviors of JM133 med-1(+) and JM134 med-1(−) are essentially indistinguishable (data not shown). (B) Histogram showing the distribution of MH33 staining intensity (classified as 0, +, ++) within arrested embryos produced by strain JM134 med-1(−) (open bars) or control strain JM133 med-1(+) (shaded bars). Images [from strain JM134 med-1(−)] represent Z-projections of deconvolved image stacks taken for each embryo; blue, DAPI staining; red, MH33 immunofluorescence. A well-formed endotube is obvious in embryos classified as “++”; a rudimentary endotube can be detected in embryos classified as “+” (see arrow). This experiment was repeated five times, scoring a total of 305 and 386 arrested embryos for strains JM133 med-1(+) and JM134 med-1(−), respectively; error bars represent standard deviations. DAPI-stained images from these experiments were used to estimate the total number of nuclei in the arrested embryos (Table 1).
Figure 5.
Microscopic images (birefringence optics) of typical arrested embryos produced by (A) strain JM135 med-1(+) and (B) strain JM136 med-1(−). This experiment was repeated a total of eight times for each genotype, scoring a total of 753 and 2162 arrested embryos for strain JM135 med-1(+) and strain JM136 med-1(−), respectively. As described in the text, >97% of arrested embryos produced by either strain still express birefringent gut granules. Bar, 100 μm.
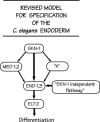
Figure 6.
Revised proposal for the transcription factor cascade responsible for specifying the C. elegans endoderm. As described in the text, we suggest that the SKN-1-activated med-1 and med-2 genes may play a relatively minor role in specifying the C. elegans endoderm. We suggest that the major pathway for endoderm specification could involve the SKN-1 transcription factor itself directly activating the end-1 and end-3 genes, as originally proposed by Z
hu
et al. (1997) or acting through an unknown factor, depicted as X. Finally, there must be a significant SKN-1 independent pathway to specify endoderm (∼30% under the current experimental conditions).
Similar articles
- Maternal deployment of the embryonic SKN-1-->MED-1,2 cell specification pathway in C. elegans.
Maduro MF, Broitman-Maduro G, Mengarelli I, Rothman JH. Maduro MF, et al. Dev Biol. 2007 Jan 15;301(2):590-601. doi: 10.1016/j.ydbio.2006.08.029. Epub 2006 Aug 22. Dev Biol. 2007. PMID: 16979152 - Neither maternal nor zygotic med-1/med-2 genes play a major role in specifying the Caenorhabditis elegans endoderm.
Captan VV, Goszczynski B, McGhee JD. Captan VV, et al. Genetics. 2007 Feb;175(2):969-74. doi: 10.1534/genetics.106.066662. Epub 2006 Dec 6. Genetics. 2007. PMID: 17151237 Free PMC article. - The Wnt effector POP-1 and the PAL-1/Caudal homeoprotein collaborate with SKN-1 to activate C. elegans endoderm development.
Maduro MF, Kasmir JJ, Zhu J, Rothman JH. Maduro MF, et al. Dev Biol. 2005 Sep 15;285(2):510-23. doi: 10.1016/j.ydbio.2005.06.022. Dev Biol. 2005. PMID: 16084508 - The Caenorhabditis elegans intestine.
McGhee JD. McGhee JD. Wiley Interdiscip Rev Dev Biol. 2013 May-Jun;2(3):347-67. doi: 10.1002/wdev.93. Epub 2012 Oct 9. Wiley Interdiscip Rev Dev Biol. 2013. PMID: 23799580 Review. - GATA factors as key regulatory molecules in the development of Drosophila endoderm.
Murakami R, Okumura T, Uchiyama H. Murakami R, et al. Dev Growth Differ. 2005 Dec;47(9):581-9. doi: 10.1111/j.1440-169X.2005.00836.x. Dev Growth Differ. 2005. PMID: 16316403 Review.
Cited by
- Roles of the Wnt effector POP-1/TCF in the C. elegans endomesoderm specification gene network.
Owraghi M, Broitman-Maduro G, Luu T, Roberson H, Maduro MF. Owraghi M, et al. Dev Biol. 2010 Apr 15;340(2):209-21. doi: 10.1016/j.ydbio.2009.09.042. Epub 2009 Oct 7. Dev Biol. 2010. PMID: 19818340 Free PMC article. - The NK-2 class homeodomain factor CEH-51 and the T-box factor TBX-35 have overlapping function in C. elegans mesoderm development.
Broitman-Maduro G, Owraghi M, Hung WW, Kuntz S, Sternberg PW, Maduro MF. Broitman-Maduro G, et al. Development. 2009 Aug;136(16):2735-46. doi: 10.1242/dev.038307. Epub 2009 Jul 15. Development. 2009. PMID: 19605496 Free PMC article. - Knockdown of SKN-1 and the Wnt effector TCF/POP-1 reveals differences in endomesoderm specification in C. briggsae as compared with C. elegans.
Lin KT, Broitman-Maduro G, Hung WW, Cervantes S, Maduro MF. Lin KT, et al. Dev Biol. 2009 Jan 1;325(1):296-306. doi: 10.1016/j.ydbio.2008.10.001. Epub 2008 Oct 19. Dev Biol. 2009. PMID: 18977344 Free PMC article. - Structure and evolution of the C. elegans embryonic endomesoderm network.
Maduro MF. Maduro MF. Biochim Biophys Acta. 2009 Apr;1789(4):250-60. doi: 10.1016/j.bbagrm.2008.07.013. Epub 2008 Aug 6. Biochim Biophys Acta. 2009. PMID: 18778800 Free PMC article. Review. - Meta-analysis of Caenorhabditis elegans single-cell developmental data reveals multi-frequency oscillation in gene activation.
Hutchison LAD, Berger B, Kohane IS. Hutchison LAD, et al. Bioinformatics. 2020 Jul 1;36(13):4047-4057. doi: 10.1093/bioinformatics/btz864. Bioinformatics. 2020. PMID: 31860066 Free PMC article.
References
- Baugh, L. R., A. A. Hill, D. K. Slonim, E. L. Brown and C. P. Hunter, 2003. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130: 889–900. - PubMed
- Baugh, L. R., A. A. Hill, J. M. Claggett, K. Hill-Harfe, J. C. Wen et al., 2005. The homeodomain protein PAL-1 specifies a lineage-specific regulatory network in the C. elegans embryo. Development 132: 1843–1854. - PubMed
- Bossinger, O., T. Fukushige, M. Claeys, G. Borgonie and J. D. McGhee, 2004. The apical disposition of the Caenorhabditis elegans intestinal terminal web is maintained by LET-413. Dev. Biol. 268: 448–456. - PubMed
- Bowerman, B., B. A. Eaton and J. R. Priess, 1992. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell 68: 1061–1075. - PubMed
- Bowerman, B., B. W. Draper, C. C. Mello and J. R. Priess, 1993. The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell 74: 443–452. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources