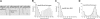Polymerization of fibrin: specificity, strength, and stability of knob-hole interactions studied at the single-molecule level - PubMed (original) (raw)
Polymerization of fibrin: specificity, strength, and stability of knob-hole interactions studied at the single-molecule level
Rustem I Litvinov et al. Blood. 2005.
Abstract
Using laser tweezers, we measured for the first time the forces of individual knob-into-hole interactions underlying fibrin polymerization. Exposure of A-knobs in desA-fibrin or its fragment from the central part of the molecule (N-terminal disulphide knot, NDSK) resulted in strong interactions with fibrinogen or fragment D (containing only a- and b-holes), producing a binding strength of approximately 125 to 130 pN. The interactions were not present in the absence of either knobs or holes and were abrogated by a specific inhibitor of fibrin polymerization, a peptide mimic of the A-knob (GPRPam). Exposure of both the A- and B-knobs in desAB-fibrin or desAB-NDSK did not change the rupture force spectra compared with the desA molecules, and their interactions with fibrinogen remained highly sensitive to GPRPam but not to GHRPam (B-knob), suggesting that neither A:b nor B:b nor B:a contacts contributed significantly to binding strength in addition to A:a contacts. The A:a interactions had a relatively small zero-force off-rate of approximately 10(-4) s(-1) and tight knob-to-hole contacts characterized by a transition state distance of approximately 0.3 nm. The results demonstrate that the knob-hole binding during thrombin-induced fibrin polymerization is driven by strong, stable, and highly specific A:a bonding, whereas A:b, B:b, or B:a interactions were not detected.
Figures
Figure 1.
Schematic representation of fibrin molecules bearing knobs and holes that bind each other in the course of fibrin polymerization. (A) A fibrin monomer is 45 nm long and consists of 3 parts, namely 2 D-regions and 1 E-region. The D-regions contain the distal portions of the coiled-coil and the C-terminal β- and γ-modules. The E-region contains the central N-terminal part of the molecule and the proximal portions of both sets of coiled-coils. The E-region has 2 pairs of binding sites named A- and B-knobs that are exposed after cleavage of FpA and FpB by thrombin. The D-regions have constitutively open a- and b-holes located in the γ- and β-modules, respectively. (B) The driving force of fibrin polymerization is the complementary binding of the A-knobs and a-holes and perhaps of the B-knobs and b-holes resulting in formation of a half-staggered 2-strand protofibril. The cartoon is based on the crystallographic data and represents approximately the relative positions and dimensions of the molecular parts. The A- and B-knobs are highly flexible and hence have not been visualized in the crystal structure to date. (C) Cartoons of the molecules used in this study (fibrinogen, fragment D, and 3 types of the fragment called N-terminal disulphide knot, NDSK). The gray and black circles represent fibrinopeptides A (FpA) and B (FpB), respectively.
Figure 2.
Rupture forces of individual protein molecules registered as multiple calibrated signals and arranged into force distribution histograms. (A) Data trace of 9 successive signals produced during repeated contacts of a desAB-fibrin–coated pedestal and fibrinogen-coated latex bead. At the moment of contact the laser trap exerts a small positive, compressive force on the pedestal and the bead. When the pedestal and the bead bind, either specifically or nonspecifically, the force on the bead increases in the negative direction until the pedestal-bead bond is ruptured, and the force rapidly returns to zero. If no attachment occurs, there is no negative rupture force. (B) DesAB-fibrin–fibrinogen rupture forces displayed as a normalized force distribution with 2 force regimes bordering at about 50 pN. The total number of contacts (n = 10 865) is taken to be 100%. (C) A control histogram of exponentially decreasing rupture forces 0 to 40 pN produced by nonspecifically interacting desAB-fibrin–and BSA-coated surfaces (n = 3689). (D) A sample of rupture force distribution of desAB-fibrin–fibrinogen interactions fitted to the Bell function combined with exponential decay for nonspecific forces less than 40 pN (n = 1378). Signals that appeared as forces below 10 pN were considered nonbinding events or zero.
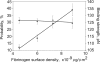
Figure 3.
DesA-NDSK/fibrinogen binding probability and strength plotted against the fibrinogen surface density. The fibrinogen surface densities were nonsaturating in this range, and the binding probabilities (•) increased linearly, whereas the binding strengths (▴) remained unchanged. Error bars indicate standard deviation.
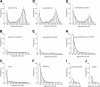
Figure 4.
The panel of rupture force spectra demonstrating the interactions of desA-fibrin and desA-NDSK with fibrinogen and fragment D along with control data for nonspecific protein-protein binding. (A,D,G) Pure interactions between desA-fibrin and fibrinogen, desA-NDSK and fibrinogen, desA-NDSK and fragment D, respectively; (B,E,H) the same interactions in the presence of 1 mM GPRPam; (C,F,I,J) negative controls with one or both interacting surfaces coated with the proteins lacking complementary D/E binding sites: fibrinogen versus fibrinogen (C), NDSK versus fibrinogen (F), desA-NDSK versus BSA (I), fibrinogen versus BSA (J). Error bars indicate standard deviation.
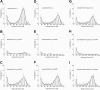
Figure 5.
The panel of rupture force spectra showing effects of the GPRPam and GHRPam peptides on the interactions of desAB-fibrin and desAB-NDSK with fibrinogen and fragment D. (A,D,G) Pure interactions between desAB-fibrin and fibrinogen, desAB-NDSK and fibrinogen, desAB-NDSK and fragment D, respectively; (B,E,H) the same interactions in the presence of 1 mM GPRPam; (C,F,I) the same interactions in the presence of 1 mM GHRPam. Error bars indicate standard deviation.
Figure 6.
A stereo figure showing the residues GPR, corresponding to the fibrin α-chain N-terminus, bound to the polymerization pocket in the globular γ-chain portion of fragment D and depicting the actual bond lengths (Å). The GPR sequence of the GPRPam peptide mimics the working part of A-knob composed of the amino acid residues (Aα17-19) exposed after cleavage of FpA. The a-hole is represented by 3 selected amino acid residues (γ329, γ330, and γ364) that are directly involved in the interaction., Considering GPRP the “surrogate A-knob” it is likely that the driving force of the A:a binding are the multiple electrostatic and hydrogen bonds between the α-amino group of AαGly17 and the guanidinium group of AαArg19 of an A-knob, on the one hand, and γAsp330 and γAsp364 along with carboxyamide of γGln329 of an a-hole, on the other hand. This aggregate of bonds stabilizing the A:a coupling is abruptly broken as soon as the knob and the hole are pulled apart to a distance of about 3 Å.
Similar articles
- Role of 'B-b' knob-hole interactions in fibrin binding to adsorbed fibrinogen.
Geer CB, Tripathy A, Schoenfisch MH, Lord ST, Gorkun OV. Geer CB, et al. J Thromb Haemost. 2007 Dec;5(12):2344-51. doi: 10.1111/j.1538-7836.2007.02774.x. Epub 2007 Sep 24. J Thromb Haemost. 2007. PMID: 17892530 - Polymerization of fibrin: Direct observation and quantification of individual B:b knob-hole interactions.
Litvinov RI, Gorkun OV, Galanakis DK, Yakovlev S, Medved L, Shuman H, Weisel JW. Litvinov RI, et al. Blood. 2007 Jan 1;109(1):130-8. doi: 10.1182/blood-2006-07-033910. Epub 2006 Aug 29. Blood. 2007. PMID: 16940416 Free PMC article. - Interactions mediated by the N-terminus of fibrinogen's Bbeta chain.
Gorkun OV, Litvinov RI, Veklich YI, Weisel JW. Gorkun OV, et al. Biochemistry. 2006 Dec 12;45(49):14843-52. doi: 10.1021/bi061430q. Biochemistry. 2006. PMID: 17144678 - The structure and biological features of fibrinogen and fibrin.
Mosesson MW, Siebenlist KR, Meh DA. Mosesson MW, et al. Ann N Y Acad Sci. 2001;936:11-30. doi: 10.1111/j.1749-6632.2001.tb03491.x. Ann N Y Acad Sci. 2001. PMID: 11460466 Review. - Fibrinogen and fibrin polymerization: appraisal of the binding events that accompany fibrin generation and fibrin clot assembly.
Mosesson MW. Mosesson MW. Blood Coagul Fibrinolysis. 1997 Jul;8(5):257-67. Blood Coagul Fibrinolysis. 1997. PMID: 9282789 Review.
Cited by
- Type I Collagen-Fibrin Mixed Hydrogels: Preparation, Properties and Biomedical Applications.
Coradin T, Wang K, Law T, Trichet L. Coradin T, et al. Gels. 2020 Oct 20;6(4):36. doi: 10.3390/gels6040036. Gels. 2020. PMID: 33092154 Free PMC article. Review. - Fibrin clots are equilibrium polymers that can be remodeled without proteolytic digestion.
Chernysh IN, Nagaswami C, Purohit PK, Weisel JW. Chernysh IN, et al. Sci Rep. 2012;2:879. doi: 10.1038/srep00879. Epub 2012 Nov 20. Sci Rep. 2012. PMID: 23170200 Free PMC article. - Mechanisms of fibrin polymerization and clinical implications.
Weisel JW, Litvinov RI. Weisel JW, et al. Blood. 2013 Mar 7;121(10):1712-9. doi: 10.1182/blood-2012-09-306639. Epub 2013 Jan 10. Blood. 2013. PMID: 23305734 Free PMC article. Review. - Development of self-assembling mixed protein micelles with temperature-modulated avidities.
Soon AS, Smith MH, Herman ES, Lyon LA, Barker TH. Soon AS, et al. Adv Healthc Mater. 2013 Jul;2(7):1045-55. doi: 10.1002/adhm.201200330. Epub 2013 Feb 26. Adv Healthc Mater. 2013. PMID: 23441099 Free PMC article. - Regulation of fibrinogen synthesis.
Dobson DA, Fish RJ, de Vries PS, Morrison AC, Neerman-Arbez M, Wolberg AS. Dobson DA, et al. Thromb Res. 2024 Oct;242:109134. doi: 10.1016/j.thromres.2024.109134. Epub 2024 Aug 28. Thromb Res. 2024. PMID: 39216273 Review.
References
- Spraggon G, Everse SJ, Doolittle RF. Crystal structures of fragment D from human fibrinogen and its crosslinked counterpart from fibrin. Nature. 1997;389: 455-462. - PubMed
- Doolittle RF. Fibrinogen and fibrin. In: Bloom AL, Thomas DP, eds. Haemostasis and Thrombosis. New York, NY: Churchill Livingstone; 1981: 163-191.
- Weisel JW. Fibrinogen and fibrin. Adv Protein Chem. 2005;70: 247-299. - PubMed
- Weisel JW, Veklich Y, Gorkun O. The sequence of cleavage of fibrinopeptides from fibrinogen is important for protofibril formation and enhancement of lateral aggregation in fibrin clots. J Mol Biol. 1993;232: 285-297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources