Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis - PubMed (original) (raw)
Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis
Takaaki Sato et al. Appl Environ Microbiol. 2005 Jul.
Abstract
We have recently developed a gene disruption system for the hyperthermophilic archaeon Thermococcus kodakaraensis by utilizing a pyrF-deficient mutant, KU25, as a host strain and the pyrF gene as a selectable marker. To achieve multiple genetic manipulations for more advanced functional analyses of genes in vivo, it is necessary to establish multiple host-marker systems or to develop a system in which repeated utilization of one marker gene is possible. In this study, we first constructed a new host strain, KU216 (DeltapyrF), by specific and almost complete deletion of endogenous pyrF through homologous recombination. In this refined host, there is no need to consider unknown mutations caused by random mutagenesis, and unlike in the previous host, KU25, there is little, if any, possibility that unintended recombination between the marker gene and the chromosomal allele occurs. Furthermore, a new host-marker combination of a trpE deletant, KW128 (DeltapyrF DeltatrpE::pyrF), and the trpE gene was developed. This system made it possible to isolate transformants through a more simple selection procedure as well as to deduce the transformation efficiency, overcoming practical disadvantages of the first system. The effects of the transformation conditions were also investigated using this system. Finally, we have also established a system in which repeated utilization of the counterselectable pyrF marker is possible through its excision by pop-out recombination. Both endogenous and exogenous sequences could be applied as tandem repeats flanking the marker pyrF for pop-out recombination. A double deletion mutant, KUW1 (DeltapyrF DeltatrpE), constructed with the pop-out strategy, was demonstrated to be a useful host for the dual markers pyrF and trpE. Likewise, a triple deletion mutant, KUWH1 (DeltapyrF DeltatrpE DeltahisD), could also be constructed. The transformation systems developed here now provide the means for extensive genetic studies in this hyperthermophilic archaeon.
Figures
FIG. 1.
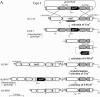
Schematic diagram of targeted disruption of pyrF, trpE, and hisD in T. kodakaraensis KOD1, KU216, and KW128 using pUDPyrF, pUDTrpE, and pUDHisD, respectively. Relevant regions of the chromosome are illustrated for (from the top) strains KOD1, KU216, KW128, and KH3. The positions of primer sets used for analyses of targeted disruption of pyrF (CHDPYR-F/CHDPYR-R, closed arrowheads), trpE (CHDTRP-R/CHDTRP-F, open arrowheads), and hisD (CHDHID-R/CHDHID-F, closed arrows) are indicated. The gray, closed, open, and striped boldface bars indicate each region spanned by the pyrF, trpE, pyrF upstream, and trpE downstream probes used in Southern blot analyses, respectively. P_pyrF_ indicates the putative promoter region of the operon containing pyrF. Black stars indicate regions of target genes that were left intact in order to avoid disturbing nearby genes as described in Materials and Methods. Gene name abbreviations: Hypo, hypothetical gene; ino1, _myo_-inositol-1-phosphate synthase; PRb, predicted RNA-binding protein; tagD, cytidylyltransferase. Restriction site abbreviations: Ap, ApaI; Hc, HincII; Hd, HindIII.
FIG. 2.
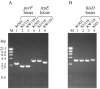
PCR analyses of T. kodakaraensis strains KU216 (Δ_pyrF_), KW128 (Δ_pyrF_ Δ_trpE_::pyrF), and KH3 (Δ_pyrF_ Δ_trpE_::pyrF Δ_hisD_::trpE). (A) Amplification of pyrF and trpE loci in strains KOD1, KU216, and KW128 using CHDPYR-R/CHDPYR-F and CHDTRP-R/CHDTRP-F as primer sets, respectively. (B) Amplification of the hisD locus in T. kodakaraensis KOD1, KU216, KW128, and KH3 using CHDHID-R and CHDHID-F as primers. Primers used for these analyses are displayed in Fig. 1. M represents the DNA size marker, HindIII-digested λ DNA.
FIG. 3.
Southern blot analyses of T. kodakaraensis strains KU216 (Δ_pyrF_), KW128 (Δ_pyrF_ Δ_trpE_::pyrF), and KH3 (Δ_pyrF_ Δ_trpE_::pyrF Δ_hisD_::trpE). (A) The pyrF upstream probe was used against genomic DNAs of KOD1 and KU216 digested with HincII. (B) The pyrF probe was used against genomic DNAs of KOD1, KU216, KW128, and KUW1 digested with ApaI. (C) The trpE downstream probe was used against genomic DNAs of KOD1, KU216, KW128, and KUW1 digested with ApaI. (D) The trpE probe was used against genomic DNAs of KOD1, KW128, and KH3 digested with HindIII. The bars on the left side of each panel indicate the mobility of fragments in the DNA size marker, HindIII-digested λ DNA. Regions spanned by probes used for these analyses are displayed in Fig. 1.
FIG. 4.
Schematic diagram of sequential disruption of trpE and hisD through excision of the pyrF marker by pop-out recombination. (A) Construction of strains KUW1 (Δ_pyrF_ Δ_trpE_) and KUWH1 (Δ_pyrF_ Δ_trpE_ Δ_hisD_) using type I pop-out vectors harboring tandem repeats of the endogenous 3′ region of the target gene flanking pyrF on both sides. (B) Construction of strains KUWc1 and KUWcHc1 using type II pop-out vectors harboring tandem repeats of the exogenous 2μ′ region flanking pyrF on both sides. The regions shaded in gray indicate the tandem repeat regions in each strategy. Open arrowheads and closed arrows indicate primer sets CHDTRP-R/CHDTRP-F and CHDHID-R/CHDHID-F for analyses of targeted disruption of trpE and hisD, respectively. Restriction site abbreviation: Ap, ApaI. All genes adjacent to the target genes are the same as those mentioned in the legend of Fig. 1.
FIG. 4.
Schematic diagram of sequential disruption of trpE and hisD through excision of the pyrF marker by pop-out recombination. (A) Construction of strains KUW1 (Δ_pyrF_ Δ_trpE_) and KUWH1 (Δ_pyrF_ Δ_trpE_ Δ_hisD_) using type I pop-out vectors harboring tandem repeats of the endogenous 3′ region of the target gene flanking pyrF on both sides. (B) Construction of strains KUWc1 and KUWcHc1 using type II pop-out vectors harboring tandem repeats of the exogenous 2μ′ region flanking pyrF on both sides. The regions shaded in gray indicate the tandem repeat regions in each strategy. Open arrowheads and closed arrows indicate primer sets CHDTRP-R/CHDTRP-F and CHDHID-R/CHDHID-F for analyses of targeted disruption of trpE and hisD, respectively. Restriction site abbreviation: Ap, ApaI. All genes adjacent to the target genes are the same as those mentioned in the legend of Fig. 1.
FIG. 5.
PCR analyses of _pyrF_-trpE double deletion mutants and _pyrF_-_trpE_-hisD triple deletion mutants of T. kodakaraensis constructed by repeated utilization of the pyrF marker using pop-out strategy. (A) Amplification of the trpE locus in strains KU216, KuW1, KUW1, KuWc1, and KUWc1 using CHDTRP-R/CHDTRP-F as a primer set. (B) Amplification of pyrF, trpE, and hisD loci in strains KU216, KUW1, and KUWH1 using CHDPYR-R/CHDPYR-F, CHDTRP-R/CHDTRP-F, and CHDHID-R/CHDHID-F as primer sets, respectively. (C) Amplification of pyrF, trpE, and hisD loci in strains KU216, KUWc1, and KUWcHc1 using CHDPYR-R/CHDPYR-F, CHDTRP-R/CHDTRP-F, and CHDHID-R/CHDHID-F as primer sets, respectively. Primer sets used for these analyses were displayed in Fig. 1 and 4. M represents the DNA size marker, HindIII-digested λ DNA.
Similar articles
- Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1.
Sato T, Fukui T, Atomi H, Imanaka T. Sato T, et al. J Bacteriol. 2003 Jan;185(1):210-20. doi: 10.1128/JB.185.1.210-220.2003. J Bacteriol. 2003. PMID: 12486058 Free PMC article. - Genetic manipulations of the hyperthermophilic piezophilic archaeon Thermococcus barophilus.
Thiel A, Michoud G, Moalic Y, Flament D, Jebbar M. Thiel A, et al. Appl Environ Microbiol. 2014 Apr;80(7):2299-306. doi: 10.1128/AEM.00084-14. Epub 2014 Jan 31. Appl Environ Microbiol. 2014. PMID: 24487541 Free PMC article. - Disruption of a sugar transporter gene cluster in a hyperthermophilic archaeon using a host-marker system based on antibiotic resistance.
Matsumi R, Manabe K, Fukui T, Atomi H, Imanaka T. Matsumi R, et al. J Bacteriol. 2007 Apr;189(7):2683-91. doi: 10.1128/JB.01692-06. Epub 2007 Jan 26. J Bacteriol. 2007. PMID: 17259314 Free PMC article. - An overview of 25 years of research on Thermococcus kodakarensis, a genetically versatile model organism for archaeal research.
Rashid N, Aslam M. Rashid N, et al. Folia Microbiol (Praha). 2020 Feb;65(1):67-78. doi: 10.1007/s12223-019-00730-2. Epub 2019 Jul 8. Folia Microbiol (Praha). 2020. PMID: 31286382 Review. - [Advance in genetic manipulation systems of hyperthermophilic archaea--a review].
Wang F, Zhang S, Huang Q, Shen Y, Ni J. Wang F, et al. Wei Sheng Wu Xue Bao. 2009 Nov;49(11):1418-23. Wei Sheng Wu Xue Bao. 2009. PMID: 20112667 Review. Chinese.
Cited by
- Genetic examination of initial amino acid oxidation and glutamate catabolism in the hyperthermophilic archaeon Thermococcus kodakarensis.
Yokooji Y, Sato T, Fujiwara S, Imanaka T, Atomi H. Yokooji Y, et al. J Bacteriol. 2013 May;195(9):1940-8. doi: 10.1128/JB.01979-12. Epub 2013 Feb 22. J Bacteriol. 2013. PMID: 23435976 Free PMC article. - Counterselection system for Geobacillus kaustophilus HTA426 through disruption of pyrF and pyrR.
Suzuki H, Murakami A, Yoshida K. Suzuki H, et al. Appl Environ Microbiol. 2012 Oct;78(20):7376-83. doi: 10.1128/AEM.01669-12. Epub 2012 Aug 10. Appl Environ Microbiol. 2012. PMID: 22885745 Free PMC article. - Polarity in archaeal operon transcription in Thermococcus kodakaraensis.
Santangelo TJ, Cubonová L, Matsumi R, Atomi H, Imanaka T, Reeve JN. Santangelo TJ, et al. J Bacteriol. 2008 Mar;190(6):2244-8. doi: 10.1128/JB.01811-07. Epub 2008 Jan 11. J Bacteriol. 2008. PMID: 18192385 Free PMC article. - Development of an efficient technique for gene deletion and allelic exchange in Geobacillus spp.
Bacon LF, Hamley-Bennett C, Danson MJ, Leak DJ. Bacon LF, et al. Microb Cell Fact. 2017 Apr 5;16(1):58. doi: 10.1186/s12934-017-0670-4. Microb Cell Fact. 2017. PMID: 28381218 Free PMC article. - The Cdc45/RecJ-like protein forms a complex with GINS and MCM, and is important for DNA replication in Thermococcus kodakarensis.
Nagata M, Ishino S, Yamagami T, Ogino H, Simons JR, Kanai T, Atomi H, Ishino Y. Nagata M, et al. Nucleic Acids Res. 2017 Oct 13;45(18):10693-10705. doi: 10.1093/nar/gkx740. Nucleic Acids Res. 2017. PMID: 28977567 Free PMC article.
References
- Aagaard, C., I. Leviev, R. N. Aravalli, P. Forterre, D. Prieur, and R. A. Garrett. 1996. General vectors for archaeal hyperthermophiles: strategies based on a mobile intron and a plasmid. FEMS Microbiol. Rev. 18:93-104. - PubMed
- Adams, M. W., and R. M. Kelly. 1998. Finding and using hyperthermophilic enzymes. Trends Biotechnol. 16:329-332. - PubMed
- Aravalli, R. N., and R. A. Garrett. 1997. Shuttle vectors for hyperthermophilic archaea. Extremophiles 1:183-191. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources