Physical limits to biochemical signaling - PubMed (original) (raw)
Physical limits to biochemical signaling
William Bialek et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Many crucial biological processes operate with surprisingly small numbers of molecules, and there is renewed interest in analyzing the impact of noise associated with these small numbers. Twenty-five years ago, Berg and Purcell showed that bacterial chemotaxis, where a single-celled organism must respond to small changes in concentration of chemicals outside the cell, is limited directly by molecule counting noise and that aspects of the bacteria's behavioral and computational strategies must be chosen to minimize the effects of this noise. Here, we revisit and generalize their arguments to estimate the physical limits to signaling processes within the cell and argue that recent experiments are consistent with performance approaching these limits.
Figures
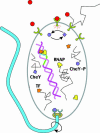
Fig. 1.
Measuring the concentration of a signaling molecule by a biological sensor, which in turn controls downstream events, is a generic task. Here, several examples are depicted schematically for E. coli. Binding of attractant/repellent molecules to a surface receptor complex modulates the rate of autophosphorylation of the associated kinase. This change in kinase activity results in a corresponding concentration change of the internal signaling molecule, CheY∼P, that controls the direction of flagellar motor rotation. Also shown is transcription initiation, where the promoter region can be regarded as a sensor for transcription factors (TF). These proteins, whose concentrations vary depending on the cell cycle and external cues, determine whether or not RNA polymerase (RNAP) turns on a gene.
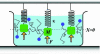
Fig. 2.
For the mass-spring system immersed in a viscous fluid, measuring the linear response of the position, X(t), to a known, small external force, F(t), determines the generalized susceptibility. From this susceptibility, the fluctuation-dissipation theorem can be used to obtain the power spectrum of fluctuations in the closed system at equilibrium, as in Eq. 11. These fluctuations provide a lower bound on the accuracy of any measurement of the position. If measurements are carried out on N identical mass-spring systems, the expected error is reduced by a factor of 1/. However, as N increases, this improvement ceases to hold, as neighboring mass-spring systems become physically close enough to experience correlated fluctuations from collisions with the particle bath, as shown. These correlations have been measured for two optically trapped colloidal particles (16).
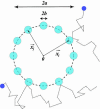
Fig. 3.
Schematic representation of a cluster of m receptors of size b, distributed uniformly on a ring of size a. For a ≫ b, the relative accuracy in measurement of the substrate concentration improves as 1/ until mb ∼ a, at which point the binding/unbinding events of nearby receptors are no longer independent.
Similar articles
- Robustness in simple biochemical networks.
Barkai N, Leibler S. Barkai N, et al. Nature. 1997 Jun 26;387(6636):913-7. doi: 10.1038/43199. Nature. 1997. PMID: 9202124 - Limits of sensing temporal concentration changes by single cells.
Mora T, Wingreen NS. Mora T, et al. Phys Rev Lett. 2010 Jun 18;104(24):248101. doi: 10.1103/PhysRevLett.104.248101. Epub 2010 Jun 14. Phys Rev Lett. 2010. PMID: 20867338 - Computer simulated evolution of a network of cell-signaling molecules.
Bray D, Lay S. Bray D, et al. Biophys J. 1994 Apr;66(4):972-7. doi: 10.1016/S0006-3495(94)80878-1. Biophys J. 1994. PMID: 8038401 Free PMC article. - Bacterial chemosensing: cooperative molecular logic.
Wolanin PM, Stock JB. Wolanin PM, et al. Curr Biol. 2004 Jun 22;14(12):R486-7. doi: 10.1016/j.cub.2004.06.018. Curr Biol. 2004. PMID: 15203024 Review. - Bioengineering models of cell signaling.
Asthagiri AR, Lauffenburger DA. Asthagiri AR, et al. Annu Rev Biomed Eng. 2000;2:31-53. doi: 10.1146/annurev.bioeng.2.1.31. Annu Rev Biomed Eng. 2000. PMID: 11701506 Review.
Cited by
- Energetic costs of cellular computation.
Mehta P, Schwab DJ. Mehta P, et al. Proc Natl Acad Sci U S A. 2012 Oct 30;109(44):17978-82. doi: 10.1073/pnas.1207814109. Epub 2012 Oct 8. Proc Natl Acad Sci U S A. 2012. PMID: 23045633 Free PMC article. - Strategies for cellular decision-making.
Perkins TJ, Swain PS. Perkins TJ, et al. Mol Syst Biol. 2009;5:326. doi: 10.1038/msb.2009.83. Epub 2009 Nov 17. Mol Syst Biol. 2009. PMID: 19920811 Free PMC article. Review. - Physical limits on bacterial navigation in dynamic environments.
Hein AM, Brumley DR, Carrara F, Stocker R, Levin SA. Hein AM, et al. J R Soc Interface. 2016 Jan;13(114):20150844. doi: 10.1098/rsif.2015.0844. J R Soc Interface. 2016. PMID: 26763331 Free PMC article. - Memory improves precision of cell sensing in fluctuating environments.
Aquino G, Tweedy L, Heinrich D, Endres RG. Aquino G, et al. Sci Rep. 2014 Jul 14;4:5688. doi: 10.1038/srep05688. Sci Rep. 2014. PMID: 25023459 Free PMC article. - Fluctuations, Correlations and the Estimation of Concentrations inside Cells.
Pérez Ipiña E, Ponce Dawson S. Pérez Ipiña E, et al. PLoS One. 2016 Mar 10;11(3):e0151132. doi: 10.1371/journal.pone.0151132. eCollection 2016. PLoS One. 2016. PMID: 26962863 Free PMC article.
References
- Schrödinger, E. (1944) What is Life? (Cambridge Univ. Press, Cambridge, U.K.).
- Rieke, F. & Baylor, D. A. (1998) Rev. Mod. Phys. 70 1027-1036.
- Boeckh, J., Kaissling, K. E. & Schneider, D. (1965) Cold Spring Harbor Symp. Quant. Biol. 30 263-281. - PubMed
- Barlow, H. B. (1981) Proc. R. Soc. London B 212 1-34. - PubMed
- Rieke, F., Warland, D., de Ruyter van Stevenick, R. & Bialek, W. (1997) Spikes: Exploring the Neural Code (MIT Press, Cambridge, MA).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources