Prediction of water and metal binding sites and their affinities by using the Fold-X force field - PubMed (original) (raw)
Comparative Study
. 2005 Jul 19;102(29):10147-52.
doi: 10.1073/pnas.0501980102. Epub 2005 Jul 8.
Affiliations
- PMID: 16006526
- PMCID: PMC1177371
- DOI: 10.1073/pnas.0501980102
Comparative Study
Prediction of water and metal binding sites and their affinities by using the Fold-X force field
Joost W H Schymkowitz et al. Proc Natl Acad Sci U S A. 2005.
Abstract
The empirical force field Fold-X was developed previously to allow rapid free energy calculations in proteins. Here, we present an enhanced version of the force field allowing prediction of the position of structural water molecules and metal ions, together called single atom ligands. Fold-X picks up 76% of water molecules found to interact with two or more polar atoms of proteins in high-resolution crystal structures and predicts their position to within 0.8 A on average. The prediction of metal ion-binding sites have success rates between 90% and 97% depending on the metal, with an overall standard deviation on the position of binding of 0.3-0.6 A. The following metals were included in the force field: Mg2+, Ca2+, Zn2+, Mn2+, and Cu2+. As a result, the current version of Fold-X can accurately decorate a protein structure with biologically important ions and water molecules. Additionally, the free energy of binding of Ca2+ and Zn2+ (i.e., the natural logarithm of the dissociation constant) and its dependence on ionic strength correlate reasonably well with the experimental data available in the literature, allowing one to discriminate between high- and low-affinity binding sites. Importantly, the accuracy of the energy prediction presented here is sufficient to efficiently discriminate between Mg2+, Ca2+, and Zn2+ binding.
Figures
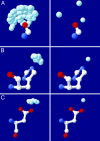
Fig. 1.
Examples of the extraction of canonical water from high-resolution structures. (Left) The “clouds” extracted from PDB files are shown. (Right) The canonical positions used for the prediction algorithm are shown. (A) Water molecules coordinated by the backbone carbonyl. (B) Zn2+ ions coordinated by the histidine Nε2 atom. (C)Mg2+ ions coordinated by aspartate Oδ1.
Fig. 2.
Examples of predicted water shells. (A) Oligopeptide binding protein (PDB ID code 1jet). (B) Hydrogenase maturation protein (PDB ID code 1gxu).
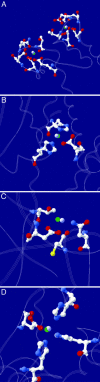
Fig. 3.
Comparison between some predicted (gray) and crystallographic (green) water- and metal-binding sites. (A)Ca2+ binding in calmodulin (PDB ID code 1cll). (B) Zn2+ binding in thermolysin (PDB ID code 8tln). (C) Structural water-binding site in RNase T1 (PDB ID code 1biv). (D)Mn2+ binding of human mitochondrial superoxide dismutase (PDB ID code 1ap6).
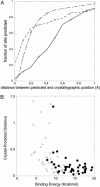
Fig. 4.
Performance of the prediction of the binding positions of metal ions. (A) The percentage of metal ions predicted within a given distance of the crystallographic position for Ca (squares), Zn (diamonds), and Cu (circles). (B) Plot of predicted binding energy vs. the distance between crystallographic and predicted positions reveals that on average highest-affinity binding sites are predicted with greatest precision.
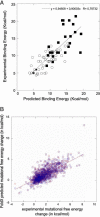
Fig. 5.
Free energy predictions. (A) Predicted free energy of binding of metal ions vs. the experimental free energy of binding (kcal/mol-1). (B) Predicted free energy of side-chain truncation of a set of 1,200 mutations vs. experimental measurements (kcal/mol-1), including predicted water (blue) molecules and without (red).
Similar articles
- Remarkable affinity and selectivity for Cs+ and uranyl (UO22+) binding to the manganese site of the apo-water oxidation complex of photosystem II.
Ananyev GM, Murphy A, Abe Y, Dismukes GC. Ananyev GM, et al. Biochemistry. 1999 Jun 1;38(22):7200-9. doi: 10.1021/bi990023u. Biochemistry. 1999. PMID: 10353831 - Interaction of different metal ions with carboxylic acid group: a quantitative study.
Bala T, Prasad BL, Sastry M, Kahaly MU, Waghmare UV. Bala T, et al. J Phys Chem A. 2007 Jul 19;111(28):6183-90. doi: 10.1021/jp067906x. Epub 2007 Jun 23. J Phys Chem A. 2007. PMID: 17585841 - Force fields including charge transfer and local polarization effects: Application to proteins containing multi/heavy metal ions.
Sakharov DV, Lim C. Sakharov DV, et al. J Comput Chem. 2009 Jan 30;30(2):191-202. doi: 10.1002/jcc.21048. J Comput Chem. 2009. PMID: 18566982
Cited by
- Construction of a stability landscape of the CH3 domain of human IgG1 by combining directed evolution with high throughput sequencing.
Traxlmayr MW, Hasenhindl C, Hackl M, Stadlmayr G, Rybka JD, Borth N, Grillari J, Rüker F, Obinger C. Traxlmayr MW, et al. J Mol Biol. 2012 Oct 26;423(3):397-412. doi: 10.1016/j.jmb.2012.07.017. Epub 2012 Jul 27. J Mol Biol. 2012. PMID: 22846908 Free PMC article. - Functional validation of hydrophobic adaptation to physiological temperature in the small heat shock protein αA-crystallin.
Posner M, Kiss AJ, Skiba J, Drossman A, Dolinska MB, Hejtmancik JF, Sergeev YV. Posner M, et al. PLoS One. 2012;7(3):e34438. doi: 10.1371/journal.pone.0034438. Epub 2012 Mar 29. PLoS One. 2012. PMID: 22479631 Free PMC article. - On human disease-causing amino acid variants: statistical study of sequence and structural patterns.
Petukh M, Kucukkal TG, Alexov E. Petukh M, et al. Hum Mutat. 2015 May;36(5):524-534. doi: 10.1002/humu.22770. Epub 2015 Apr 6. Hum Mutat. 2015. PMID: 25689729 Free PMC article. - Computer-aided antibody design.
Kuroda D, Shirai H, Jacobson MP, Nakamura H. Kuroda D, et al. Protein Eng Des Sel. 2012 Oct;25(10):507-21. doi: 10.1093/protein/gzs024. Epub 2012 Jun 2. Protein Eng Des Sel. 2012. PMID: 22661385 Free PMC article. Review. - The Advances and Limitations of the Determination and Applications of Water Structure in Molecular Engineering.
Zsidó BZ, Bayarsaikhan B, Börzsei R, Szél V, Mohos V, Hetényi C. Zsidó BZ, et al. Int J Mol Sci. 2023 Jul 22;24(14):11784. doi: 10.3390/ijms241411784. Int J Mol Sci. 2023. PMID: 37511543 Free PMC article. Review.
References
- Mendes, J., Guerois, R. & Serrano, L. (2002) Curr. Opin. Struct. Biol. 12, 441-446. - PubMed
- Lazaridis, T. & Karplus, M. (2000) Curr. Opin. Struct. Biol. 10, 139-145. - PubMed
- Guerois, R., Nielsen, J. E. & Serrano, L. (2002) J. Mol. Biol. 320, 369-387. - PubMed
- Fernandez-Ballester, G., Blanes-Mira, C. & Serrano, L. (2004) J. Mol. Biol. 335, 619-629. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous