JunD activates transcription of the human ferritin H gene through an antioxidant response element during oxidative stress - PubMed (original) (raw)
JunD activates transcription of the human ferritin H gene through an antioxidant response element during oxidative stress
Yoshiaki Tsuji. Oncogene. 2005.
Abstract
Ferritin is the major intracellular iron storage protein that sequesters excess free iron to minimize generation of iron-catalysed reactive oxygen species. We previously demonstrated that expression of ferritin heavy chain (ferritin H) was induced by pro-oxidants, which is a part of cellular antioxidant response to protect cells from oxidative damage. In this study, we have identified that the antioxidant/electrophile response element (ARE) located 4.5 kb upstream to the human ferritin H transcription initiation site is responsible for the oxidant response. The human ferritin H ARE comprises two copies of bidirectional AP1 motifs. Mutations in each AP1 motif significantly impaired protein binding and the function of the ARE, indicating that both of the AP1 motifs are required for pro-oxidant-mediated activation of the ferritin H gene. We identified that JunD, an AP1 family basic-leucine zipper (bZip) transcription factor, is one of the ferritin H ARE binding proteins and activates ferritin H transcription in HepG2 hepatocarcinoma cells. Gel retardation assay demonstrated that H2O2 (hydrogen peroxide) or t-BHQ (tert-butylhydroquinone) treatment increased total protein binding as well as JunD binding to the ferritin H ARE. Chromatin immunoprecipitation assay showed that H2O2 treatment induced JunD binding to the ferritin H ARE. Both H2O2 and t-BHQ induced phosphorylation of JunD at Ser-100, an activated form of JunD. Furthermore, overexpression of JunD induced endogenous ferritin H protein synthesis. Since JunD has recently been demonstrated to protect cells from several stress stimuli including oxidative stress, these results suggest that, in addition to NFE2-related factor 2 (Nrf2) as a major ARE regulatory protein, JunD is another ARE regulatory protein for transcriptional activation of the human ferritin H gene and probably other antioxidant genes containing the conserved ARE sequences by which JunD may confer cytoprotection during oxidative stress.
Figures
Figure 1
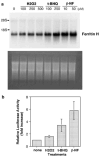
H2O2, t-BHQ and _β_-NF induce ferritin H mRNA at transcriptional level. (a) HepG2 cells were treated with indicated concentrations of H2O2, t-BHQ or _β_-NF for 24 h and 10 _μ_g of total RNA from each treatment was subjected to hybridization with a ferritin H cDNA probe (top). Equal amounts of RNA loading and transfer to nitrocellulose membrane were verified by ethidium bromide staining (bottom). (b) A measure of 1 _μ_g of −5.2 kb ferritin H-luciferase plasmid was transiently transfected into HepG2 cells by calcium phosphate method, followed by treatment with 250 μ
m
H2O2, 250 μ
m
t-BHQ or 50 μ
m
_β_-NF for 24 h. Luciferase activity with no treatment was defined as 1.0, and the results from four independent experiments and s.e.'s are shown
Figure 2
The human ferritin H gene is regulated by the proximal and a far-upstream enhancer element, and the latter is responsible for transcriptional activation by oxidative stressors. A measure of 1 _μ_g of a human ferritin H-luciferase reporter construct (−5.2, −4.0, −0.15, −0.03 kb or zero) was transiently transfected into HepG2 cells, followed by treatment with 250 μ
m
H2O2, 250 μ
m
t-BHQ or 50 μ
m
_β_-NF for 24 h and luciferase assays. Luciferase activity from HepG2 cells transfected with the −0.15 kb construct without treatment was defined as 1.0. The results from six experiments and standard errors are shown
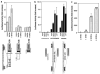
Figure 3
A human ferritin H ARE. (a) The 4.5 kb-upstream enhancer of the human ferritin H gene contains composite AP1 motifs. A measure of 1 _μ_g of a human ferritin H-luciferase reporter construct (−4.5, −4.4 or −0.15 kb) was transiently transfected into HepG2 cells, followed by treatment with 250 μ
m
H2O2 or 250 μ
m
t-BHQ for 24 h and luciferase assays. Luciferase activity from HepG2 cells transfected with the −4.4 kb construct without treatment was defined as 1.0. The results from four experiments and s.e.'s are shown. (b) The composite AP1 motifs serve as a basal enhancer and are sufficient to activate transcription of the ferritin H gene in response to oxidative stressors. One copy of a double-stranded oligonucleotide containing the composite AP1 motifs was inserted into −0.03 kb TATA-ferritin H luciferase construct in sense or antisense orientation, and 1 _μ_g of each luciferase construct was transiently transfected into HepG2 cells. The transfected HepG2 cells were treated with 250 μ
m
H2O2, 250 μ
m
t-BHQ or 50 μ
m
_β_-NF for 24 h followed by luciferase assays. Luciferase activity from HepG2 cells transfected with the antisense insertion construct without treatment was defined as 1.0. The results from three experiments and standard errors are shown. (c) Multiple copies of the oligonucleotides containing the composite AP1 motifs were inserted into −0.03 kb TATA-ferritin H luciferase construct in antisense orientation, and 1 _μ_g of each luciferase construct was transiently transfected into HepG2 cells and luciferase assays were carried out. Luciferase activity from HepG2 cells transfected with no insertion construct was defined as 1.0. The results from three experiments and standard errors are shown
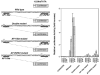
Figure 4
Both AP1-like and AP1/NFE2 sites are essential for the functional ARE. One copy of a double-stranded oligonucleotide containing wt AP1-like and AP1/NFE2 motifs, point mutations in either AP1-like site or AP1/NFE2 site, or mutations in both sites was inserted into a TATA-ferritin H luciferase construct, and 1 _μ_g of each luciferase construct was transiently transfected into HepG2 cells. The transfected HepG2 cells were treated with 250 μ
m
H2O2, 250 μ
m
t-BHQ or 50 μ
m
_β_-NF for 24 h followed by luciferase assays. Luciferase activity from HepG2 cells transfected with the wt insertion construct and no treatment was defined as 1.0. The results from four experiments and s.e.'s are shown
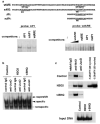
Figure 5
JunD is one of ferritin H ARE binding proteins. (a) Gel retardation assays were carried out by incubating 20 _μ_g of nuclear extracts isolated from growing HepG2 cells with 32P-labeled, double-stranded AP1/NFE2 probe (left) or wt ARE probe (right) in the presence of 40- to 50-fold molar excess of the indicated double-stranded competitor oligonucleotides. Nucleotide sequences of oligonucleotides used in the assays are shown on top with mutations denoted by asterisks. (b) HepG2 cells were treated with 100 μ
m
H2O2 or 100 μ
m
t-BHQ for 4 h followed by isolation of nuclear extracts. 20 _μ_g of HepG2 nuclear extracts were incubated with either rabbit IgG or anti-JunD antibody (rabbit) for 2 h at 4°C prior to the addition of a 32P-labeled AP1/NFE2 probe. A supershift band induced by anti-JunD antibody was indicated by an arrow. (c) HepG2 cells were treated with 100 μ
m
H2O2 or 100 μ
m
t-BHQ for 6 h and ChIP assay was carried out with rabbit IgG, anti-Nrf2 or anti-JunD antibody. Immunoprecipitated DNA, input DNA from each treatment (approximately 1/20 of cell lysate used for each immunoprecipitation) or 1 pg of pBluescript −5.2 kb ferritin H luciferase DNA was used for detection of 155 bp ferritin H ARE in 28 cycles of PCR reaction in addition to 30 cycles of PCR with pBluescript −5.2 kb ferritin H luciferase DNA for confirmation of a linear range of the DNA amplification
Figure 6
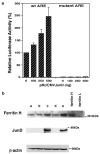
JunD activates the human ferritin H ARE. (a) A measure of 1 _μ_g of pBluescript wtARE ferritin H luciferase or mARE ferritin H luciferase (wtARE and mARE DNA sequences are shown in Figure 5a) was cotransfected with indicated amounts of pRc/CMVJunD into HepG2 cells. Total input of plasmid DNA was equalized to 2 _μ_g by adding pRc/CMV empty vector DNA and 0.1 _μ_g of pRL-CMV. Cells were harvested for dual luciferase assays 48–60 h after DNA transfection. DNA transfection was performed in duplicate, and the luciferase expression after normalization with pRL-CMV from five independent experiments are shown with s.e.'s. The luciferase activity in cell lysates cotransfected with wtARE ferritin H luciferase and pRc/CMV empty vector was defined as 100%. (b) A total of 7 × 106 K562 cells were transiently transfected with no DNA (lane a), 15 _μ_g of pRc/CMV (lane b), pRc/CMVJunD (lane c), pcDNAINeoJunD (lane d) or pCMVJunD (lane e) by electroporation and were incubated for 65 h. Cell lysates (50 _μ_g) were subjected to Western blotting for anti-ferritin H, anti-JunD and anti-_β_-actin. JunD cDNA cloned into pcDNAINeo vector was expected to express JunD protein but no expression for an uncertain reason
Figure 7
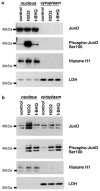
JunD expression in nucleus and phosphorylation at Ser100 during oxidative stress. (a) HepG2 cells were treated with 100 μ
m
H2O2 or 100 μ
m
t-BHQ for 1.5 h, (b) K562 cells transiently transfected with 20 _μ_g of pRc/CMVJunD were incubated for 45 h, followed by treatment with 100 μ
m
H2O2 or 100 μ
m
t-BHQ for 4 h. In both (a) and (b), cells were harvested for isolation of nuclear and cytoplasmic extracts, and 30 _μ_g of proteins were subjected to Western blotting for anti-JunD or anti-phospho-cJun (Ser73), which also detects phospho-JunD at Ser100. The purity of nuclear and cytoplasmic fractions was assessed by Western blotting with anti-histone H1 and anti-lactose dehydrogenase (LDH), respectively
Similar articles
- Hemin-mediated regulation of an antioxidant-responsive element of the human ferritin H gene and role of Ref-1 during erythroid differentiation of K562 cells.
Iwasaki K, Mackenzie EL, Hailemariam K, Sakamoto K, Tsuji Y. Iwasaki K, et al. Mol Cell Biol. 2006 Apr;26(7):2845-56. doi: 10.1128/MCB.26.7.2845-2856.2006. Mol Cell Biol. 2006. PMID: 16537925 Free PMC article. - FER-1, an enhancer of the ferritin H gene and a target of E1A-mediated transcriptional repression.
Tsuji Y, Akebi N, Lam TK, Nakabeppu Y, Torti SV, Torti FM. Tsuji Y, et al. Mol Cell Biol. 1995 Sep;15(9):5152-64. doi: 10.1128/MCB.15.9.5152. Mol Cell Biol. 1995. PMID: 7651432 Free PMC article. - JunD reduces tumor angiogenesis by protecting cells from oxidative stress.
Gerald D, Berra E, Frapart YM, Chan DA, Giaccia AJ, Mansuy D, Pouysségur J, Yaniv M, Mechta-Grigoriou F. Gerald D, et al. Cell. 2004 Sep 17;118(6):781-94. doi: 10.1016/j.cell.2004.08.025. Cell. 2004. PMID: 15369676 - Nrf2/ARE regulated antioxidant gene expression in endothelial and smooth muscle cells in oxidative stress: implications for atherosclerosis and preeclampsia.
Mann GE, Niehueser-Saran J, Watson A, Gao L, Ishii T, de Winter P, Siow RC. Mann GE, et al. Sheng Li Xue Bao. 2007 Apr 25;59(2):117-27. Sheng Li Xue Bao. 2007. PMID: 17437032 Review. - The many faces of the octahedral ferritin protein.
Watt RK. Watt RK. Biometals. 2011 Jun;24(3):489-500. doi: 10.1007/s10534-011-9415-8. Epub 2011 Jan 26. Biometals. 2011. PMID: 21267633 Review.
Cited by
- Multiple functions of the histone chaperone Jun dimerization protein 2.
Tsai MH, Wuputra K, Lin YC, Lin CS, Yokoyama KK. Tsai MH, et al. Gene. 2016 Sep 30;590(2):193-200. doi: 10.1016/j.gene.2016.03.048. Epub 2016 Apr 8. Gene. 2016. PMID: 27041241 Free PMC article. Review. - Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling.
Ray PD, Huang BW, Tsuji Y. Ray PD, et al. Cell Signal. 2012 May;24(5):981-90. doi: 10.1016/j.cellsig.2012.01.008. Epub 2012 Jan 20. Cell Signal. 2012. PMID: 22286106 Free PMC article. Review. - Stress-activated cap'n'collar transcription factors in aging and human disease.
Sykiotis GP, Bohmann D. Sykiotis GP, et al. Sci Signal. 2010 Mar 9;3(112):re3. doi: 10.1126/scisignal.3112re3. Sci Signal. 2010. PMID: 20215646 Free PMC article. Review. - Aldo-keto reductase 1 family B7 is the gene induced in response to oxidative stress in the livers of Long-Evans Cinnamon rats.
Jia G, Takahashi R, Zhang Z, Tsuji Y, Sone H. Jia G, et al. Int J Oncol. 2006 Oct;29(4):829-38. Int J Oncol. 2006. PMID: 16964378 Free PMC article. - Solving the Problem of Assessing Synergy and Antagonism for Non-Traditional Dosing Curve Compounds Using the DE/ZI Method: Application to Nrf2 Activators.
Repash EM, Pensabene KM, Palenchar PM, Eggler AL. Repash EM, et al. Front Pharmacol. 2021 Jun 7;12:686201. doi: 10.3389/fphar.2021.686201. eCollection 2021. Front Pharmacol. 2021. PMID: 34163365 Free PMC article.
References
- Ainbinder E, Bergelson S, Pinkus R, Daniel V. Eur J Biochem. 1997;243:49–57. - PubMed
- Arosio P, Levi S. Free Radic‘ Biol Med. 2002;33:457–463. - PubMed
- Beaumont C, Jain SK, Bogard M, Nordmann Y, Drysdale J. J Biol Chem. 1987;262:10619–10623. - PubMed
- Beaumont C, Seyhan A, Yachou AK, Grandchamp B, Jones R. J Biol Chem. 1994;269:20281–20288. - PubMed
- Bevilacqua MA, Faniello MC, Quaresima B, Tiano MT, Giuliano P, Feliciello A, Avvedimento VE, Cimino F, Costanzo F. J Biol Chem. 1997;272:20736–20741. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK060007/DK/NIDDK NIH HHS/United States
- R01 DK060007-01A1/DK/NIDDK NIH HHS/United States
- R01 DK060007-02/DK/NIDDK NIH HHS/United States
- DK-60007/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases