Identification of critical active-site residues in angiotensin-converting enzyme-2 (ACE2) by site-directed mutagenesis - PubMed (original) (raw)
Identification of critical active-site residues in angiotensin-converting enzyme-2 (ACE2) by site-directed mutagenesis
Jodie L Guy et al. FEBS J. 2005 Jul.
Abstract
Angiotensin-converting enzyme-2 (ACE2) may play an important role in cardiorenal disease and it has also been implicated as a cellular receptor for the severe acute respiratory syndrome (SARS) virus. The ACE2 active-site model and its crystal structure, which was solved recently, highlighted key differences between ACE2 and its counterpart angiotensin-converting enzyme (ACE), which are responsible for their differing substrate and inhibitor sensitivities. In this study the role of ACE2 active-site residues was explored by site-directed mutagenesis. Arg273 was found to be critical for substrate binding such that its replacement causes enzyme activity to be abolished. Although both His505 and His345 are involved in catalysis, it is His345 and not His505 that acts as the hydrogen bond donor/acceptor in the formation of the tetrahedral peptide intermediate. The difference in chloride sensitivity between ACE2 and ACE was investigated, and the absence of a second chloride-binding site (CL2) in ACE2 confirmed. Thus ACE2 has only one chloride-binding site (CL1) whereas ACE has two sites. This is the first study to address the differences that exist between ACE2 and ACE at the molecular level. The results can be applied to future studies aimed at unravelling the role of ACE2, relative to ACE, in vivo.
Figures
Figure 1
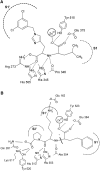
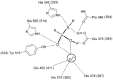
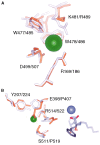
Schematic view of the active site of ACE2 and tACE. Binding interactions of the inhibitor (A) MLN‐4760 at the active site of ACE2 and (B) lisinopril at the active site of tACE. Hydrogen bonds to the ligand are shown (dotted lines). The different binding subsites are labelled. Adapted from [17].
Figure 2
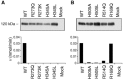
Expression of soluble ACE2 mutants. Medium, taken from mock‐transfected (empty vector) HEK293 cells and HEK293 cells transiently expressing soluble ACE2, was concentrated in a 10‐kDa cut‐off column. Aliquots, containing 30 µg total protein, were separated by SDS/PAGE (6% polyacrylamide gel) and then analysed by immunoelectrophoretic blotting using a human ACE2 polyclonal antibody (top panel). Total protein (30 µg) was incubated with the ACE2‐specific fluorogenic peptide, Mca‐APK(Dnp) (25 µ
m
), as described in Experimental Procedures. Enzyme activity is expressed as mol product formed per min (bottom panel). Values are the mean of duplicate determinations.
Figure 3
Role of His505 and His345 in catalysis. Schematic of the proposed reaction intermediate of ACE2, showing the importance of His345 and His505. Hydrogen bonds to the ligand are shown (dotted lines).The equivalent residues in tACE are given in parentheses.
Figure 4
Effect of chloride ions on the activity of the ACE2 mutants (R169Q/R514Q). Medium, taken from HEK293 cells stably expressing soluble ACE2, was concentrated in a 10‐kDa cut‐off column and extensively dialysed against 50 m
m
Hepes/KOH buffer, pH 7.5, to remove chloride ions. Total protein (10 µg) was incubated with the ACE2‐specific fluorogenic peptide, Mca‐APK(Dnp) (25 µ
m
), as described in Experimental Procedures in the absence (grey) or presence (black) of NaCl (500 m
m
). Enzyme activity is expressed as mol product formed over time. Product was quantified using pure standards. Values are the mean of four independent determinations.
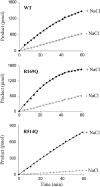
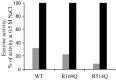
Figure 5
Activities of wild‐type and R169Q and R514Q ACE2 mutants in the absence (grey) and presence (black) of NaCl (500 m
m
). Medium, taken from HEK293 cells stably expressing soluble ACE2, was concentrated in a 10‐kDa cut‐off column and extensively dialysed against 50 m
m
Hepes/KOH buffer, pH 7.5, to remove chloride ions. Total protein (10 µg) was incubated with the ACE2‐specific fluorogenic peptide, Mca‐APK(Dnp) (25 µ
m
), as described in Experimental Procedures in the absence (grey bars) or presence (black bars) of NaCl (500 m
m
). Enzyme activity (mol product formed·min−1) is expressed as the percentage of activity with 500 m
m
NaCl. Product was quantified using pure standards. Values are mean ± SE from four independent determinations.
Figure 6
Chloride binding to ACE2 (yellow) and tACE (white). (A) Binding site of CL1 in ACE2 and tACE; (B) binding site of CL2 in ACE2 and tACE. Residue numbering for ACE2 is first. The chloride ion is green and the zinc ion is grey (both in spacefill). (B) The lisinopril ligand is coloured according to atom type (CPK) and the chloride ion is shown with a reduced radius to demonstrate its overlap with Glu398 in ACE2 more clearly.
Similar articles
- Residues affecting the chloride regulation and substrate selectivity of the angiotensin-converting enzymes (ACE and ACE2) identified by site-directed mutagenesis.
Rushworth CA, Guy JL, Turner AJ. Rushworth CA, et al. FEBS J. 2008 Dec;275(23):6033-42. doi: 10.1111/j.1742-4658.2008.06733.x. FEBS J. 2008. PMID: 19021774 Free PMC article. - Angiotensin-converting enzyme-2 (ACE2): comparative modeling of the active site, specificity requirements, and chloride dependence.
Guy JL, Jackson RM, Acharya KR, Sturrock ED, Hooper NM, Turner AJ. Guy JL, et al. Biochemistry. 2003 Nov 18;42(45):13185-92. doi: 10.1021/bi035268s. Biochemistry. 2003. PMID: 14609329 - ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis.
Towler P, Staker B, Prasad SG, Menon S, Tang J, Parsons T, Ryan D, Fisher M, Williams D, Dales NA, Patane MA, Pantoliano MW. Towler P, et al. J Biol Chem. 2004 Apr 23;279(17):17996-8007. doi: 10.1074/jbc.M311191200. Epub 2004 Jan 30. J Biol Chem. 2004. PMID: 14754895 Free PMC article. - Angiotensin-converting enzyme-2: a molecular and cellular perspective.
Warner FJ, Smith AI, Hooper NM, Turner AJ. Warner FJ, et al. Cell Mol Life Sci. 2004 Nov;61(21):2704-13. doi: 10.1007/s00018-004-4240-7. Cell Mol Life Sci. 2004. PMID: 15549171 Free PMC article. Review. - Membrane-associated zinc peptidase families: comparing ACE and ACE2.
Guy JL, Lambert DW, Warner FJ, Hooper NM, Turner AJ. Guy JL, et al. Biochim Biophys Acta. 2005 Aug 1;1751(1):2-8. doi: 10.1016/j.bbapap.2004.10.010. Epub 2004 Nov 6. Biochim Biophys Acta. 2005. PMID: 16054014 Free PMC article. Review.
Cited by
- Exploring the disruption of SARS-CoV-2 RBD binding to hACE2.
Carter C, Airas J, Gladden H, Miller BR 3rd, Parish CA. Carter C, et al. Front Chem. 2023 Oct 24;11:1276760. doi: 10.3389/fchem.2023.1276760. eCollection 2023. Front Chem. 2023. PMID: 37954960 Free PMC article. - In silico ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19.
Vardhan S, Sahoo SK. Vardhan S, et al. Comput Biol Med. 2020 Sep;124:103936. doi: 10.1016/j.compbiomed.2020.103936. Epub 2020 Jul 28. Comput Biol Med. 2020. PMID: 32738628 Free PMC article. - Angiotensin-converting enzyme 2 and angiotensin 1-7: novel therapeutic targets.
Jiang F, Yang J, Zhang Y, Dong M, Wang S, Zhang Q, Liu FF, Zhang K, Zhang C. Jiang F, et al. Nat Rev Cardiol. 2014 Jul;11(7):413-26. doi: 10.1038/nrcardio.2014.59. Epub 2014 Apr 29. Nat Rev Cardiol. 2014. PMID: 24776703 Free PMC article. Review. - Vectored immunoprophylaxis and treatment of SARS-CoV-2 infection in a preclinical model.
Tada T, Minnee J, Landau NR. Tada T, et al. Proc Natl Acad Sci U S A. 2023 Jun 6;120(23):e2303509120. doi: 10.1073/pnas.2303509120. Epub 2023 May 30. Proc Natl Acad Sci U S A. 2023. PMID: 37252952 Free PMC article. - Heteromeric Solute Carriers: Function, Structure, Pathology and Pharmacology.
Fairweather SJ, Shah N, Brӧer S. Fairweather SJ, et al. Adv Exp Med Biol. 2021;21:13-127. doi: 10.1007/5584_2020_584. Adv Exp Med Biol. 2021. PMID: 33052588 Review.
References
- Tipnis SR, Hooper NM, Hyde R, Karran E, Christie G & Turner AJ (2000) A human homolog of angiotensin‐converting enzyme: cloning and functional expression as a captopril‐insensitive carboxypeptidase. J Biol Chem 275, 33238–33243. - PubMed
- Donoghue M, Hsieh F, Baronas E, Godbout K, Gosselin M, Stagliano N, Donovan M, Woolf B, Robison K, Jeyaseelan R et al. (2000) A novel angiotensin‐converting enzyme‐related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ Res 87, E1–E9. - PubMed
- Vickers C, Hales P, Kaushik V, Dick L, Gavin J, Tang J, Godbout K, Parsons T, Baronas E, Hsieh F et al. (2002) Hydrolysis of biological peptides by human angiotensin‐converting enzyme‐related carboxypeptidase. J Biol Chem 277, 14838–14843. - PubMed
- Crackower MA, Sarao R, Oudit GY, Yagil C, Kozieradzki I, Scanga SE, Oliveira‐dos‐Santos AJ, da Costa J, Zhang L, Pei Y et al. (2002) Angiotensin‐converting enzyme 2 is an essential regulator of heart function. Nature 417, 822–828. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous