Midbodies and phragmoplasts: analogous structures involved in cytokinesis - PubMed (original) (raw)
Review
Midbodies and phragmoplasts: analogous structures involved in cytokinesis
Marisa S Otegui et al. Trends Cell Biol. 2005 Aug.
Erratum in
- Trends Cell Biol. 2005 Oct;15(10):517
Abstract
Cytokinesis is an event common to all organisms that involves the precise coordination of independent pathways involved in cell-cycle regulation and microtubule, membrane, actin and organelle dynamics. In animal cells, the spindle midzone/midbody with associated endo-membrane system are required for late cytokinesis events, including furrow ingression and scission. In plants, cytokinesis is mediated by the phragmoplast, an array of microtubules, actin filaments and associated molecules that act as a framework for the future cell wall. In this article (which is part of the Cytokinesis series), we discuss recent studies that highlight the increasing number of similarities in the components and function of the spindle midzone/midbody in animals and the phragmoplast in plants, suggesting that they might be analogous structures.
Figures
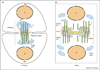
Figure 1
Overview of animal and plant cytokinesis. (a) Cytokinesis in animal cells. The spindle midzone/midbody forms when microtubules (MTs) from opposite poles overlap. It consists of the overlapping microtubules as well as associated proteins that bundle these MTs and other proteins that together form a dense protein matrix. This matrix excludes antibodies against MTs, giving a stereotypical region devoid of staining. As the furrow ingresses, the midzone is swept into one larger structure called the midbody. The Golgi and endoplasmic reticulum (ER) membranes are also found in the midbody during telophase to cytokinesis. It is proposed that vesicles (V) traffic along the midbody microtubules toward the ingressing furrow. (b) Cytokinesis in somatic plant cells. The forming cell plate is assisted by the phragmoplast at the future site of the new cell wall. Two topographic regions can be distinguished in the phragmoplast: the phragmoplast midline (Ph M), where the opposing set of microtubules interdigitate, and the distal phragmoplast (distal Ph), at both sides of the phragmoplast midline. A filamentous cell-plate assembly matrix (CPAM) accumulates at the phragmoplast midline. Key: MT, microtubule (green); N, nucleus (tan); V, vesicle (yellow); Golgi (pale blue); midbody matrix (gray box); CPAM (gray circles).
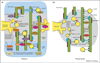
Figure 2
Models for assembly of the midbody and phragmoplast. (a) Model depicting midbody organization. Overlapping microtubule plus-ends are embedded within the midbody matrix. The midbody microtubules are stabilized and crosslinked by several kinesins, microtubule-associated proteins (MAPs), kinases and other proteins, such as MKLP1/ZEN-4/PAV, MgcRACGAP/CYK-4, PRC1/SPD-1/FEO/Ase1p, chTOG/ZYG-9/Msps, TACC/TAC-1, CLIP-170, EB1, KLP61F/BimC, and the chromosomal passengers Aurora B/AIR-2, Survivin/BIR-1, INCENP/ICP-1, -2 and Borealin/CSC-1. PRC1 is possibly transported by KIF4 along the midzone microtubules (MTs). It is possible that KIF4 also transports Golgi-derived vesicles along midbody MTs where they fuse with the ingressing furrow. ZEN-4 and PRC1 are phosphorylated by Cdk1. Golgi assembly occurs when Cdk1 activity is low in telophase. Golgi stack assembly is also promoted by the degradation of Polo kinase and MEK1 in telophase (U: ubiquitin moiety). NIR2, a Golgi-associated protein, is found more prominently on the cleavage furrow as well as the spindle midzone in telophase to cytokinesis in a complex with Polo kinase. Membrane fusion occurs at the tips and along the ingressing furrow and involves a variety of proteins such as the exocyst complex, SNAREs, dynamin/DYN-1 and their interactors. Note: to simplify the figure, we have omitted several homologs/orthologs of several proteins. (b) Model depicting phragmoplast organization in plant cells. Microtubule plus-ends enclosed inside the cell-plate assembly matrix (CPAM) are stabilized and crosslinked by several MAPs, such as MAP55-1, MAP65-3, MOR1/GEM1, and by kinesin motor proteins. Golgi-derived vesicles are transported to the division plane along MTs by a kinesin motor protein, where they fuse with others while enclosed by the CPAM. Membrane fusion at the cell plate involves a variety of proteins such as tethering factors, SNAREs and their interactors. Formation of tubular domains in the developing cell plate appears to be mediated by DRP1A.
Similar articles
- Centralspindlin links the mitotic spindle to the plasma membrane during cytokinesis.
Lekomtsev S, Su KC, Pye VE, Blight K, Sundaramoorthy S, Takaki T, Collinson LM, Cherepanov P, Divecha N, Petronczki M. Lekomtsev S, et al. Nature. 2012 Dec 13;492(7428):276-9. doi: 10.1038/nature11773. Nature. 2012. PMID: 23235882 - KIF4 regulates midzone length during cytokinesis.
Hu CK, Coughlin M, Field CM, Mitchison TJ. Hu CK, et al. Curr Biol. 2011 May 24;21(10):815-24. doi: 10.1016/j.cub.2011.04.019. Epub 2011 May 12. Curr Biol. 2011. PMID: 21565503 Free PMC article. - Katanin p60 contributes to microtubule instability around the midbody and facilitates cytokinesis in rat cells.
Matsuo M, Shimodaira T, Kasama T, Hata Y, Echigo A, Okabe M, Arai K, Makino Y, Niwa S, Saya H, Kishimoto T. Matsuo M, et al. PLoS One. 2013 Nov 26;8(11):e80392. doi: 10.1371/journal.pone.0080392. eCollection 2013. PLoS One. 2013. PMID: 24303010 Free PMC article. - Phosphoregulation of Microtubule Assembly and Disassembly for Phragmoplast Expansion During Plant Cytokinesis.
Lee YJ, Liu B. Lee YJ, et al. Bioessays. 2025 May;47(5):e202500004. doi: 10.1002/bies.202500004. Epub 2025 Mar 3. Bioessays. 2025. PMID: 40025940 Review. - MAP65: a bridge linking a MAP kinase to microtubule turnover.
Sasabe M, Machida Y. Sasabe M, et al. Curr Opin Plant Biol. 2006 Dec;9(6):563-70. doi: 10.1016/j.pbi.2006.09.010. Epub 2006 Sep 29. Curr Opin Plant Biol. 2006. PMID: 17011227 Review.
Cited by
- Dynamic actin gene family evolution in primates.
Zhu L, Zhang Y, Hu Y, Wen T, Wang Q. Zhu L, et al. Biomed Res Int. 2013;2013:630803. doi: 10.1155/2013/630803. Epub 2013 Jun 6. Biomed Res Int. 2013. PMID: 23841080 Free PMC article. - Resurrecting remnants: the lives of post-mitotic midbodies.
Chen CT, Ettinger AW, Huttner WB, Doxsey SJ. Chen CT, et al. Trends Cell Biol. 2013 Mar;23(3):118-28. doi: 10.1016/j.tcb.2012.10.012. Epub 2012 Dec 11. Trends Cell Biol. 2013. PMID: 23245592 Free PMC article. Review. - Rab-A2 and Rab-A3 GTPases define a trans-golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate.
Chow CM, Neto H, Foucart C, Moore I. Chow CM, et al. Plant Cell. 2008 Jan;20(1):101-23. doi: 10.1105/tpc.107.052001. Epub 2008 Jan 31. Plant Cell. 2008. PMID: 18239134 Free PMC article. - The Arabidopsis exocyst complex is involved in cytokinesis and cell plate maturation.
Fendrych M, Synek L, Pecenková T, Toupalová H, Cole R, Drdová E, Nebesárová J, Sedinová M, Hála M, Fowler JE, Zársky V. Fendrych M, et al. Plant Cell. 2010 Sep;22(9):3053-65. doi: 10.1105/tpc.110.074351. Epub 2010 Sep 24. Plant Cell. 2010. PMID: 20870962 Free PMC article. - Phosphorylation of NtMAP65-1 by a MAP kinase down-regulates its activity of microtubule bundling and stimulates progression of cytokinesis of tobacco cells.
Sasabe M, Soyano T, Takahashi Y, Sonobe S, Igarashi H, Itoh TJ, Hidaka M, Machida Y. Sasabe M, et al. Genes Dev. 2006 Apr 15;20(8):1004-14. doi: 10.1101/gad.1408106. Epub 2006 Apr 5. Genes Dev. 2006. PMID: 16598040 Free PMC article.
References
- Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. - PubMed
- Straight AF, et al. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–1747. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources