Gene expression and antiviral activity of alpha/beta interferons and interleukin-29 in virus-infected human myeloid dendritic cells - PubMed (original) (raw)
Comparative Study
Gene expression and antiviral activity of alpha/beta interferons and interleukin-29 in virus-infected human myeloid dendritic cells
Pamela Osterlund et al. J Virol. 2005 Aug.
Abstract
Dendritic cells (DCs) respond to microbial infections by undergoing phenotypic maturation and by producing multiple cytokines. In the present study, we analyzed the ability of influenza A and Sendai viruses to induce DC maturation and activate tumor necrosis factor alpha (TNF-alpha), alpha/beta interferon (IFN-alpha/beta), and IFN-like interleukin-28A/B (IFN-lambda2/3) and IL-29 (IFN-lambda1) gene expression in human monocyte-derived myeloid DCs (mDC). The ability of influenza A virus to induce mDC maturation or enhance the expression of TNF-alpha, IFN-alpha/beta, interleukin-28 (IL-28), and IL-29 genes was limited, whereas Sendai virus efficiently induced mDC maturation and enhanced cytokine gene expression. Influenza A virus-induced expression of TNF-alpha, IFN-alpha, IFN-beta, IL-28, and IL-29 genes was, however, dramatically enhanced when cells were pretreated with IFN-alpha. IFN-alpha priming led to increased expression of Toll-like receptor 3 (TLR3), TLR7, TLR8, MyD88, TRIF, and IFN regulatory factor 7 (IRF7) genes and enhanced influenza-induced phosphorylation and DNA binding of IRF3. Influenza A virus also enhanced the binding of NF-kappaB to the respective NF-kappaB elements of the promoters of IFN-beta and IL-29 genes. In mDC IL-29 induced MxA protein expression and possessed antiviral activity against influenza A virus, although this activity was lower than that of IFN-alpha or IFN-beta. Our results show that in human mDCs viruses can readily induce the expression of IL-28 and IL-29 genes whose gene products are likely to contribute to the host antiviral response.
Figures
FIG. 1.
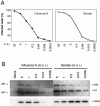
Infectivity of influenza A and Sendai viruses in human monocyte-derived mDCs. (A) Monocyte-derived mDCs were infected with influenza A/Beijing/353/89 H3N2 or Sendai (Cantell strain) viruses with different virus doses as indicated in the figure. Virus-infected cells were fixed with 1% paraformaldehyde for 15 min, permeabilized with 0.1% Triton X-100 for 5 min, and stained with whole virus-specific antibodies and FITC-labeled secondary antibodies, followed by FACS analysis. The results are the means of two donor mDCs. (B) mDCs from three separate donors were infected with different viral doses. Cells were collected and pooled, and protein samples (10 μg/lane) were separated by SDS-PAGE, followed by Western blot analysis with virus and MxA protein-specific antibodies. Virus doses, viral proteins, and MxA protein as indicated in the figure. m.o.i., MOI.
FIG. 2.
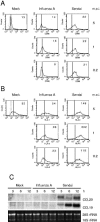
Maturation of influenza A and Sendai virus-infected mDCs. (A and B) mDCs from three separate blood donors were infected with different doses of influenza A or Sendai viruses. After 18 h of virus infection mDCs were collected, pooled, and fixed with 1% paraformaldehyde. The cells were stained, and the expression of CD86 (panel A) and HLA class II (panel B) was analyzed by FACS. Values within the insets represent the geometric mean fluorescence intensities. Dotted lines indicate respective isotype controls. The results from one representative experiment of three are shown. (C) Virus-induced expression of CCL20 and CCL19 mRNAs. mDCs obtained from three blood donors were infected with influenza A virus or Sendai virus at an MOI of 5. Cells were collected at different times as indicated in the figure, and the total cellular RNA was collected and analyzed by Northern blotting with CCL20 and CCL19 cDNA probes. Ethidium bromide staining was used to control equal sample loading.
FIG. 3.
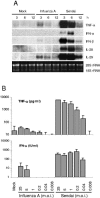
Virus-induced expression of TNF-α, IFN-α/β, IL-28, and IL-29 genes. (A) mDCs from three blood donors were infected with influenza A (MOI = 5) or Sendai (MOI = 5) viruses for different periods of time. Cells were collected, total cellular RNA was isolated and processed for Northern blotting. The filters were probed with TNF-α, IFN-α (IFN-α1 probe), IFN-β, IL-28, and IL-29 specific cDNA probes. Ethidium bromide staining was used to control equal sample loading. (B) Virus dose-dependent production of TNF-α and IFN-α/β. mDCs were infected with different doses of influenza A or Sendai viruses for 20 h, and the cell culture supernatants were collected. TNF-α levels were measured by ELISA, and IFN-α/β levels were determined by a biological IFN assay. The results are the means (±1 standard deviation) of four different experiments, each performed with cells from three blood donors. Virus doses (MOI) as indicated in the figure.
FIG. 4.
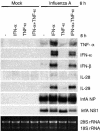
Effect of IFN-α and TNF-α priming on influenza A virus-induced cytokine gene expression. mDCs were pretreated with IFN-α (100 IU/ml), TNF-α (5 ng/ml), or both for 6 h, followed by infection with influenza A virus (MOI = 5) for 6 h. Total cellular RNA was collected and analyzed by Northern blotting for TNF-α, IFN-α (IFN-α1 probe), IFN-β, IL-28, IL-29, and influenza A virus NP and NS1 mRNA expression. Ethidium bromide staining was used to control equal sample loading.
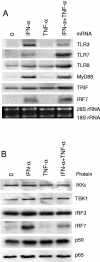
FIG. 5.
Expression of cytokine-specific signaling molecules in DC. (A) Monocyte-derived DCs were stimulated with IFN-α (100 IU/ml) and/or TNF-α (5 ng/ml) for 6 h, and total cellular RNA was isolated. The expression of TLR3, TLR7, TLR8, MyD88, TRIF, and IRF7 genes was analyzed by Northern blotting. Ethidium bromide staining was used to control equal sample loading. (B) The protein levels of IKKɛ, TBK1, IRF3, IRF7, p50, and p65 in IFN-α (100 IU/ml)- and/or TNF-α (5 ng/ml)-stimulated mDCs (16 h stimulation) was analyzed by Western blotting.
FIG. 6.
Effect of IFN-α and TNF-α priming on influenza A virus-induced IRF3, IRF7, and NF-κB DNA binding to the ISRE and NF-κB sites of IFN-α, IFN-β, and IL-29 promoters. mDCs were pretreated with IFN-α (100 IU/ml), TNF-α (5 ng/ml), or both for 16 h, followed by infection with influenza A virus (MOI = 5) for 6 h. Cells were collected, and proteins from nuclear extracts were precipitated with IFN-α (α14), IFN-β, and IL-29 ISRE and NF-κB oligonucleotides as indicated in the figure. Oligonucleotide-bound proteins were analyzed by Western blotting with anti-IRF3, anti-IRF3 S396, anti-IRF7, anti-p50, or anti-p65 antibodies as indicated in the figure.
FIG. 7.
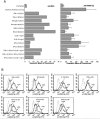
Antiviral effect of IFN-α/β, IFN-γ, and IL-29 in DCs. mDCs from four individual blood donors were pretreated alone or in various combinations (as indicated) with doses of 10 or 100 IU of natural leukocyte IFN-α (IFN-α)/ml, recombinant IFN-α2b, recombinant IFN-β, or natural leukocyte IFN-γ and 1 or 10 ng of recombinant IL-29/ml for 24 h, followed by influenza A virus infection (MOI = 1) for 18 h. (A) Prior to virus infection part of the cells was collected, pooled, fixed, and permeabilized with 0.1% Triton X-100 for 5 min, followed by staining with anti-MxA and FITC-labeled secondary antibodies. MxA protein expression is shown as geometric mean fluorescence intensities. IFN- or IL-29-pretreated and virus-infected cells were collected separately, fixed, and stained for cell surface expression of influenza A virus glycoproteins. The results are the means (±1 standard deviation) from four different donors. IFN and IL-29 doses were as indicated in the figure. (B) FACS analysis of the expression of viral proteins on the surface of IFN- or IL-29-pretreated cells. Dotted lines represent uninfected cells. Solid lines represent influenza A virus glycoprotein-specific expression in virus-infected (InfA) or in cytokine-pretreated and virus-infected cells (cytokine + InfA) as indicated in the figure. The results from the cells of one representative blood donor (out of four) are shown.
Similar articles
- IFN-alpha regulates TLR-dependent gene expression of IFN-alpha, IFN-beta, IL-28, and IL-29.
Sirén J, Pirhonen J, Julkunen I, Matikainen S. Sirén J, et al. J Immunol. 2005 Feb 15;174(4):1932-7. doi: 10.4049/jimmunol.174.4.1932. J Immunol. 2005. PMID: 15699120 - Toll-like receptor 3 mediates a more potent antiviral response than Toll-like receptor 4.
Doyle SE, O'Connell R, Vaidya SA, Chow EK, Yee K, Cheng G. Doyle SE, et al. J Immunol. 2003 Apr 1;170(7):3565-71. doi: 10.4049/jimmunol.170.7.3565. J Immunol. 2003. PMID: 12646618 - Early alpha/beta interferon production by myeloid dendritic cells in response to UV-inactivated virus requires viral entry and interferon regulatory factor 3 but not MyD88.
Hidmark AS, McInerney GM, Nordström EK, Douagi I, Werner KM, Liljeström P, Karlsson Hedestam GB. Hidmark AS, et al. J Virol. 2005 Aug;79(16):10376-85. doi: 10.1128/JVI.79.16.10376-10385.2005. J Virol. 2005. PMID: 16051830 Free PMC article. - [Toll-like receptors that sense viral infection].
Seya T, Shingai M, Matsumoto M. Seya T, et al. Uirusu. 2004 Jun;54(1):1-8. doi: 10.2222/jsv.54.1. Uirusu. 2004. PMID: 15449898 Review. Japanese. - Toll-like receptor 3: a link between toll-like receptor, interferon and viruses.
Matsumoto M, Funami K, Oshiumi H, Seya T. Matsumoto M, et al. Microbiol Immunol. 2004;48(3):147-54. doi: 10.1111/j.1348-0421.2004.tb03500.x. Microbiol Immunol. 2004. PMID: 15031527 Review.
Cited by
- Antiviral and myocyte protective effects of IL-28A in coxsackievirus B3-induced myocarditis.
Wang S, Huang X, Zhang J, Huang C. Wang S, et al. Braz J Infect Dis. 2015 Mar-Apr;19(2):132-40. doi: 10.1016/j.bjid.2014.10.007. Epub 2014 Dec 17. Braz J Infect Dis. 2015. PMID: 25528576 Free PMC article. Retracted. - Incoming influenza A virus evades early host recognition, while influenza B virus induces interferon expression directly upon entry.
Österlund P, Strengell M, Sarin LP, Poranen MM, Fagerlund R, Melén K, Julkunen I. Österlund P, et al. J Virol. 2012 Oct;86(20):11183-93. doi: 10.1128/JVI.01050-12. Epub 2012 Aug 1. J Virol. 2012. PMID: 22855501 Free PMC article. - Asian and African lineage Zika viruses show differential replication and innate immune responses in human dendritic cells and macrophages.
Österlund P, Jiang M, Westenius V, Kuivanen S, Järvi R, Kakkola L, Lundberg R, Melén K, Korva M, Avšič-Županc T, Vapalahti O, Julkunen I. Österlund P, et al. Sci Rep. 2019 Oct 31;9(1):15710. doi: 10.1038/s41598-019-52307-1. Sci Rep. 2019. PMID: 31673117 Free PMC article. - Interferon-λ in HCV Infection and Therapy.
Pagliaccetti NE, Robek MD. Pagliaccetti NE, et al. Viruses. 2010 Aug;2(8):1589-1602. doi: 10.3390/v2081589. Epub 2010 Aug 5. Viruses. 2010. PMID: 21994696 Free PMC article. - Unique type I interferon responses determine the functional fate of migratory lung dendritic cells during influenza virus infection.
Moltedo B, Li W, Yount JS, Moran TM. Moltedo B, et al. PLoS Pathog. 2011 Nov;7(11):e1002345. doi: 10.1371/journal.ppat.1002345. Epub 2011 Nov 3. PLoS Pathog. 2011. PMID: 22072965 Free PMC article.
References
- Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732-738. - PubMed
- Au, W. C., W. S. Yeow, and P. M. Pitha. 2001. Analysis of functional domains of interferon regulatory factor 7 and its association with IRF-3. Virology 280:273-282. - PubMed
- Banchereau, J., F. Briere, C. Caux, J. Davoust, S. Lebecque, Y. J. Liu, B. Pulendran, and K. Palucka. 2000. Immunobiology of dendritic cells. Annu. Rev. Immunol. 18:767-811. - PubMed
- Barchet, W., A. Krug, M. Cella, C. Newby, J. A. Fischer, A. Dzionek, A. Pekosz, and M. Colonna. 2005. Dendritic cells respond to influenza virus through TLR7- and PKR-independent pathways. Eur. J. Immunol. 35:236-242. - PubMed
- Benvenuti, F., C. Lagaudriere-Gesbert, I. Grandjean, C. Jancic, C. Hivroz, A. Trautmann, O. Lantz, and S. Amigorena. 2004. Dendritic cell maturation controls adhesion, synapse formation, and the duration of the interactions with naive T lymphocytes. J. Immunol. 172:292-301. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous