Ranks of genuine associations in whole-genome scans - PubMed (original) (raw)
Comparative Study
Ranks of genuine associations in whole-genome scans
Dmitri V Zaykin et al. Genetics. 2005 Oct.
Abstract
With the recent advances in high-throughput genotyping techniques, it is now possible to perform whole-genome association studies to fine map causal polymorphisms underlying important traits that influence susceptibility to human diseases and efficacy of drugs. Once a genome scan is completed the results can be sorted by the association statistic value. What is the probability that true positives will be encountered among the first most associated markers? When a particular polymorphism is found associated with the trait, there is a chance that it represents either a "true" or a "false" association (TA vs. FA). Setting appropriate significance thresholds has been considered to provide assurance of sufficient odds that the associations found to be significant are genuine. However, the problem with genome scans involving thousands of markers is that the statistic values of FAs can reach quite extreme magnitudes. In such situations, the distributions corresponding to TAs and the most extreme FAs become comparable and significance thresholds tend to penalize TAs and FAs in a similar fashion. When sorting between true and false associations, the "typical" place (i.e., rank) of TAs among the most significant outcomes becomes important, ordered by the association statistic value. The distribution of ranks that we study here allows calculation of several useful quantities. In particular, it gives the number of most significant markers needed for a follow-up study to guarantee that a true association is included with certain probability. This can be calculated conditionally on having applied a multiple-testing correction. Effects of multilocus (e.g., haplotype association) tests and impact of linkage disequilibrium on the distribution of ranks associated with TAs are evaluated and can be taken into account.
Figures
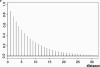
Figure 1.
Diffusion-generated _P_-value correlation decay. The distance between two neighboring markers (1 unit on the _x_-axis) is 15 kb.
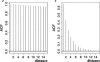
Figure 2.
_P_-value correlation decay within (left) and between (right) LD blocks. The distance between two neighboring markers (1 unit on the _x_-axis) is 15 kb.
Figure 3.
Expected power associated with individual effects (bottom line) given overall power for three effects (m = 3) (top line).
Figure 4.
Plot of degrees of freedom vs. significance level (α) for 80% power tests at the 70% quantile.
Figure 5.
Plot of degrees of freedom vs. significance level (α) for 80% power tests at the 5% quantile.
Similar articles
- Efficient multilocus association testing for whole genome association studies using localized haplotype clustering.
Browning BL, Browning SR. Browning BL, et al. Genet Epidemiol. 2007 Jul;31(5):365-75. doi: 10.1002/gepi.20216. Genet Epidemiol. 2007. PMID: 17326099 - The application of the entropy-based statistic for genomic association study of QTL.
Xiang Y, Li Y, Liu Z, Sun Z. Xiang Y, et al. J Genet Genomics. 2008 Mar;35(3):183-8. doi: 10.1016/S1673-8527(08)60025-9. J Genet Genomics. 2008. PMID: 18355762 - On selecting markers for association studies: patterns of linkage disequilibrium between two and three diallelic loci.
Garner C, Slatkin M. Garner C, et al. Genet Epidemiol. 2003 Jan;24(1):57-67. doi: 10.1002/gepi.10217. Genet Epidemiol. 2003. PMID: 12508256 Review. - Evaluating statistical significance in two-stage genomewide association studies.
Lin DY. Lin DY. Am J Hum Genet. 2006 Mar;78(3):505-9. doi: 10.1086/500812. Epub 2006 Jan 11. Am J Hum Genet. 2006. PMID: 16408254 Free PMC article. - Prospects and pitfalls in whole genome association studies.
Lawrence RW, Evans DM, Cardon LR. Lawrence RW, et al. Philos Trans R Soc Lond B Biol Sci. 2005 Aug 29;360(1460):1589-95. doi: 10.1098/rstb.2005.1689. Philos Trans R Soc Lond B Biol Sci. 2005. PMID: 16096108 Free PMC article. Review.
Cited by
- Phenotype Uniformity in Combined-Stress Environments has a Different Genetic Architecture than in Single-Stress Treatments.
Makumburage GB, Stapleton AE. Makumburage GB, et al. Front Plant Sci. 2011 May 4;2:12. doi: 10.3389/fpls.2011.00012. eCollection 2011. Front Plant Sci. 2011. PMID: 22645526 Free PMC article. - Estimating the posterior probability that genome-wide association findings are true or false.
Bukszár J, McClay JL, van den Oord EJ. Bukszár J, et al. Bioinformatics. 2009 Jul 15;25(14):1807-13. doi: 10.1093/bioinformatics/btp305. Epub 2009 May 6. Bioinformatics. 2009. PMID: 19420056 Free PMC article. - Genetics of osteoarthritis.
Aubourg G, Rice SJ, Bruce-Wootton P, Loughlin J. Aubourg G, et al. Osteoarthritis Cartilage. 2022 May;30(5):636-649. doi: 10.1016/j.joca.2021.03.002. Epub 2021 Mar 17. Osteoarthritis Cartilage. 2022. PMID: 33722698 Free PMC article. Review. - A novel method, the Variant Impact On Linkage Effect Test (VIOLET), leads to improved identification of causal variants in linkage regions.
Martin LJ, Ding L, Zhang X, Kissebah AH, Olivier M, Benson DW. Martin LJ, et al. Eur J Hum Genet. 2014 Feb;22(2):243-7. doi: 10.1038/ejhg.2013.120. Epub 2013 Jun 5. Eur J Hum Genet. 2014. PMID: 23736220 Free PMC article. - Diverse convergent evidence in the genetic analysis of complex disease: coordinating omic, informatic, and experimental evidence to better identify and validate risk factors.
Ciesielski TH, Pendergrass SA, White MJ, Kodaman N, Sobota RS, Huang M, Bartlett J, Li J, Pan Q, Gui J, Selleck SB, Amos CI, Ritchie MD, Moore JH, Williams SM. Ciesielski TH, et al. BioData Min. 2014 Jun 30;7:10. doi: 10.1186/1756-0381-7-10. eCollection 2014. BioData Min. 2014. PMID: 25071867 Free PMC article.
References
- Benjamini, Y., and Y. Hochberg,, 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57: 289–300.
- Goldstein, D. B., K. R. Ahmadi, M. E. Weale and N. W. Wood, 2003. Genome scans and candidate gene approaches in the study of common diseases and variable drug responses. Trends Genet. 19: 615–622. - PubMed
- Hamilton, D. C., and D. E. Cole, 2004. Standardizing a composite measure of linkage disequilibrium. Ann. Hum. Genet. 68: 234–239. - PubMed
- Ioannidis, J. P., E. E. Ntzani, T. A. Trikalinos and D. G. Contopoulos-Ioannidis, 2001. Replication validity of genetic association studies. Nat. Genet. 29: 306–309. - PubMed
- Kammerer, S., R. B. Roth, R. Reneland, G. Marnellos, C. R. Hoyal et al., 2004. Large-scale association study identifies ICAM gene region as breast and prostate cancer susceptibility locus. Cancer Res. 64: 8906–8910. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous