Mammalian polyhomeotic homologues Phc2 and Phc1 act in synergy to mediate polycomb repression of Hox genes - PubMed (original) (raw)
Mammalian polyhomeotic homologues Phc2 and Phc1 act in synergy to mediate polycomb repression of Hox genes
Kyo-Ichi Isono et al. Mol Cell Biol. 2005 Aug.
Erratum in
- Mol Cell Biol. 2014 Jul;34(14):2771
Abstract
The Polycomb group (PcG) gene products form multimeric protein complexes and contribute to anterior-posterior (A-P) specification via the transcriptional regulation of Hox cluster genes. The Drosophila polyhomeotic genes and their mammalian orthologues, Phc1, Phc2, and Phc3, encode nuclear proteins that are constituents of evolutionarily conserved protein complexes designated class II PcG complexes. In this study, we describe the generation and phenotypes of Phc2-deficient mice. We show posterior transformations of the axial skeleton and premature senescence of mouse embryonic fibroblasts associated with derepression of Hox cluster genes and Cdkn2a genes, respectively. Synergistic actions of a Phc2 mutation with Phc1 and Rnf110 mutations during A-P specification, coimmunoprecipitation of their products from embryonic extracts, and chromatin immunoprecipitation by anti-Phc2 monoclonal antibodies suggest that Hox repression by Phc2 is mediated through the class II PcG complexes, probably via direct binding to the Hox locus. The genetic interactions further reveal the functional overlap between Phc2 and Phc1 and a strict dose-dependent requirement during A-P specification and embryonic survival. Functional redundancy between Phc2 and Phc1 leads us to hypothesize that the overall level of polyhomeotic orthologues in nuclei is a parameter that is critical in enabling the class II PcG complexes to exert their molecular functions.
Figures
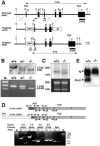
FIG. 1.
Association of Phc2 with other components of class II PcG complexes during embryogenesis. Antibodies used for IP and immunoblotting (IB) are indicated over each lane and beside each blot, respectively. Phc2 was coimmunoprecipited with Rnf110, Rnf2, and Phc1 from 11.5-dpc embryonic extracts. Original whole-cell extract was loaded on the lanes labeled “Input”; mock immunoprecipitation without primary antibody was performed as a negative control on the lanes labeled “None.”
FIG. 2.
Disruption of the Phc2 gene in mice. (A) Diagram of the Phc2 locus, the targeting vector, and the targeted allele. The PGKneo and pMC1-tk expression cassettes were used for positive and negative selection, respectively. The relevant positions of the restriction sites (EcoRI, E; XhoI, X), locations of the external probe and PCR primers, and sizes of diagnostic fragments are indicated. (B) Southern (top) and PCR (bottom) analyses for genotyping. For Southern blotting, genomic DNA was digested by EcoRI and probed with the 3′ probe, as indicated in panel A. For PCR, a mixture of three primers (p1, p2, and p3 in panel A) was used. Lane M, molecular size markers. (C) Northern analysis of Phc2 expression in 11.5-dpc wild-type and homozygous embryos (top). Ethidium bromide staining of the same gel is shown below. (D) RT-PCR analysis of Phc2 expression. The locations of the PCR primers are indicated on both the 2.5- and 3.8-kb transcripts (top). The asterisks indicate specific PCR products. Note the presence of truncated Phc2 transcripts lacking exons 2 and 3. (E) Immunoprecipitation and immunoblot analyses of Phc2 expression in 11.5-dpc embryos.
FIG. 3.
Skeletal alterations in _Phc2_−/− mice and gene dose-dependent skeletal alterations in _Phc2_-Phc1 compound mutants. (A) Lateral views of the occipitocervical region; overviews of individual C1, C2, C3, C5, C6, and C7 vertebrae; ventral views of rib cages; and overviews of scapulas. In the lateral views of the occipitocervical region, C1 and C2 are indicated numerically, and asterisks indicate the ectopic arch or piece. In Phc2+/−-_Phc1_−/− mice, a bracket indicates the segmentation of the exoccipital bone and an arrow indicates the anterior tuberculus of C1. In C1 and C2 vertebrae, closed and open arrows, respectively, indicate the anterior tuberculus and odontoid processes. In C5, C6, and C7, closed and open arrowheads, respectively, indicate the anterior tuberculus for vertebral arteries and ectopic ribs. In ventral views of the rib cages, the numbers of vertebrae to which ribs are attached are indicated. (B) Ventral views of skulls and tympanic bones. In the skull, a cleft in the secondary palate is emphasized by the use of yellow lines indicating the medial edges of the palatine. Note the absence of cartilaginous condensation at the center of the sphenoid in Phc2+/−-_Phc1_−/− mice. (C) Summary of posterior transformations. Each arrow represents the homeotic transformation of vertebrae. (a) Supraoccipital bone→C1, appearance of the ectopic arch or bone; (b) C1→C2, fusion of the odontoid process to the C1 vertebra; (c) C2→C3; (d) C5→C6, association of the anterior tuberculus with the C5 vertebra; (e) C6→T1, association of the cervical rib with the C6; (f) C7→T1, association of the cervical rib with C7; (g) C7→T2, prominent spinous process on C7; (h) T1→T2, prominent spinous process on T1; (i) T2→T3, lack of prominent spinous process in T2; (j) T6→T8, dissociation of the sixth rib from the sternum; (k) T7→T8, dissociation of the seventh rib from the sternum; (l) T13→L1, loss of the rib from the 20th vertebra; (m) L4→S1, formation of a sacroiliac joint in the 25th vertebra; (n) L6→S1, formation of a sacroiliac joint in the 26th vertebra; (o) S3→Ca1, appearance of the first caudal vertebra in the 29th vertebra; (p) S4→Ca1, appearance of the first caudal vertebra in the 30th vertebra.
FIG. 4.
Changes in Hox gene expression in _Phc2_−/− mice. Expression of Hox genes in 11.5-dpc _Phc2_−/− (A, C, E, G, I, and K) and wild-type (B, D, F, H, J, and L) embryos. The expressions of Hoxb3 (A and B), Hoxb4 (C and D), Hoxb6 (E and F), Hoxc6 (G and H), Hoxd4 (I and J), and Hoxa5 (K and L) are shown. Several prevertebrae are numbered in each panel, and dotted lines indicate the segment boundaries. The arrows indicate anterior boundaries of Hox gene expression.
FIG. 5.
Defects in the proliferation of _Phc2_−/− MEFs. (A) Cell proliferation using a 3T9 protocol. At 3-day intervals, the total numbers of wild-type and _Phc2_−/− MEFs per culture were determined. The error bars indicate standard errors of the mean. (B) Proliferation of MEFs from respective genotypes at passage 5. (C) Northern analysis of p16 and p19 ARF transcripts in wild-type and _Phc2_−/− MEFs at passages 3, 4, and 5 (P3, P4, and P5). The same gel was stained with ethidium bromide to verify the loaded amounts. (D) Distribution of Phc2 proteins in the Cdkn2a genomic region in MEFs. Representative results show the association of the Phc2 protein with the second exonic region of Cdkn2a, using the adam34 coding region as a negative control. Immunoprecipitation of the chromatin was performed using the anti-Phc2 antibody (+); mock immunoprecipitation (−) was used as a negative control. Equivalent amounts of genomic DNA from different sources were adjusted and designated ×1.
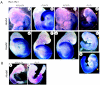
FIG. 6.
Expression of Hoxb4 and Hoxb8. (A) Hoxb4 (frames a to d) and Hoxb8 (frames e to h) expression in 9.5-dpc wild-type and Phc2 and Phc1 single homozygotes and double homozygotes; the genotypes are indicated across the top. Each specimen was subjected to chromogenic reaction for the same length of time. The specimen shown in frame h was subjected to a longer chromogenic reaction (frame hí). Dotted lines indicate the otic vesicles in this series of figures. The prospective anterior boundary of the expression is indicated by a yellow arrow in double homozygotes. All pictures for both Hoxb4 and Hoxb8 expressions were taken under the same magnification. (B) Hoxb8 expression in 8.5-dpc wild-type and double-homozygous embryos; the pictures were taken under the same magnification.
FIG. 7.
Gene dose-dependent skeletal alterations in wild-type and Phc2 and Rnf110 single homozygotes and double homozygotes. (A) The genotypes of the specimens are shown along the top of the frames. (Frames a to d) Lateral views of the occipitocervical region. The arrowheads indicate the ribs associated with the eighth vertebrae. C1 and C2 are indicated numerically, and asterisks indicate the ectopic arch or piece. (Frames d and h) An asterisk indicates an ectopic arch that represents perfect segmentation of the exoccipital bone. Red arrows indicate ectopic cartilaginous condensation bridging the hyoid bone and styloid process, while the dotted line indicates the lack of cartilaginous condensation between the occipital and sphenoid bones. (Frames e to h) Ventral views of the skull. (Frames i to l) Ventral views of rib cages. The numbers of vertebrae to which ribs are attached are indicated numerically. Asterisks indicate the sternums that are shifted anteriorly in Phc2 single (frame j) and double (frame l) homozygotes. (Frames m to p) Overviews of the scapula. (B) Summary of posterior transformations: (a) supraoccipital bone→C1, appearance of the ectopic arch or bone; (b) C1→C2, fusion of the odontoid process to the C1 vertebra; (c) C7→T1, association of the cervical rib with C7; (d) T7→T8, dissociation of the seventh rib from the sternum; (e) T13→L1, loss of the rib from the 20th vertebra; (f) L6→S1, formation of a sacroiliac joint in the 26th vertebra; (g) S4→Ca1, appearance of the first caudal vertebra in the 30th vertebra; (h) more perfect supraoccipital bone→C1 transformation, perfect segmentation of supra- and exoccipital bones; (i) C1→C3, overall structure similar to C3 rather than C2; (j) C5→C6, association of the anterior tuberculus with the C5 vertebra; (k) C6→T1, association of the cervical rib with C6; (l) T12→L1, loss of the rib from the 19th vertebra; (m) L4→S1, formation of a sacroiliac joint in the 25th vertebra; (n) S3→Ca1, appearance of the first caudal vertebra in the 29th vertebra.
FIG. 8.
Distribution of Phc2 proteins in the Hoxb8 genomic region at 11.5 dpc. (A) Representative results showing the association of the Phc2 protein with the Hoxb8 enhancer and promoter and intronic and Hoxb7 enhancer regions, with the adam34 coding region as a negative control. The positions of PCR fragments are shown in panel B. Tissues of 12.5-dpc embryos were dissected, and chromatin fractions were purified from transcriptionally silent anterior (A) and active posterior (P) tissues (mesoderm plus neurectoderm). Immunoprecipitation of the chromatin was performed by the anti-Phc2 antibody (+); mock immunoprecipitation (−) was used as a negative control. Equivalent amounts of genomic DNAs from different sources were adjusted and designated 1′. (B) The distribution of Phc2 in the Hoxb8 genomic region is schematically represented. The black and gray squares represent the degree of enrichment of immunoprecipitated genomic DNA in posterior and anterior tissues, respectively: each square represents a twofold enrichment in comparison with the unfractionated “input” DNA. The distribution of Phc1 is shown as a reference.
FIG. 9.
The expression of Phc2 and Phc1 in various tissues, lymphocyte subpopulations, and B lymphocytes induced by various extracellular stimuli. (A) The expression of Phc2 and Phc1 was examined by reverse transcriptase-PCR in various adult tissues and whole embryos at 7, 11, 15, and 17 days postcoitus. Distilled water (dH2O) and mouse genomic DNA (gDNA) were used as negative controls; β-actin was used to normalize the amounts of cDNA. (B) The expression of Phc2 and Phc1 was examined in developing B and T lymphocytes in the bone marrow and thymus, respectively, and B-cell subpopulations in naïve and immunized spleens. Mature splenic B cells were fractionated into germ center B (GCB), follicular B (FOB), marginal zone B (MZB), and newly formed B (NFB) subpopulations. Phc2 did not exhibit any variation in expression levels, in contrast to Phc1, where the level of expression was variable. (C) Expression of Phc2 and Phc1 in resting B lymphocytes upon activation by B-cell receptor engagement by α-IgM, BAFF stimulation, CD40 ligation by α-CD40, and LPS stimulation.
Similar articles
- Mammalian Polycomb complexes are required for Peyer's patch development by regulating lymphoid cell proliferation.
Sato T, Endoh M, Yoshida H, Yasuo S, Katsuno T, Saito Y, Isono K, Koseki H. Sato T, et al. Gene. 2006 Sep 1;379:166-74. doi: 10.1016/j.gene.2006.05.006. Epub 2006 May 25. Gene. 2006. PMID: 16815646 - Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression.
Akasaka T, van Lohuizen M, van der Lugt N, Mizutani-Koseki Y, Kanno M, Taniguchi M, Vidal M, Alkema M, Berns A, Koseki H. Akasaka T, et al. Development. 2001 May;128(9):1587-97. doi: 10.1242/dev.128.9.1587. Development. 2001. PMID: 11290297 - Distinct roles of Polycomb group gene products in transcriptionally repressed and active domains of Hoxb8.
Fujimura Y, Isono K, Vidal M, Endoh M, Kajita H, Mizutani-Koseki Y, Takihara Y, van Lohuizen M, Otte A, Jenuwein T, Deschamps J, Koseki H. Fujimura Y, et al. Development. 2006 Jun;133(12):2371-81. doi: 10.1242/dev.02405. Epub 2006 May 10. Development. 2006. PMID: 16687444 - [Maintenance of cellular memory by Polycomb group genes].
Netter S, Boivin A. Netter S, et al. C R Acad Sci III. 2001 Jul;324(7):577-88. doi: 10.1016/s0764-4469(01)01329-4. C R Acad Sci III. 2001. PMID: 11475999 Review. French. - [Role of mammalian polycomb group gene products in embryo genesis].
Fujimura Y, Koseki H. Fujimura Y, et al. Tanpakushitsu Kakusan Koso. 2005 May;50(6 Suppl):563-8. Tanpakushitsu Kakusan Koso. 2005. PMID: 15926481 Review. Japanese. No abstract available.
Cited by
- The roles of Polycomb repressive complexes in mammalian development and cancer.
Piunti A, Shilatifard A. Piunti A, et al. Nat Rev Mol Cell Biol. 2021 May;22(5):326-345. doi: 10.1038/s41580-021-00341-1. Epub 2021 Mar 15. Nat Rev Mol Cell Biol. 2021. PMID: 33723438 Review. - Distinct roles for CKM-Mediator in controlling Polycomb-dependent chromosomal interactions and priming genes for induction.
Dimitrova E, Feldmann A, van der Weide RH, Flach KD, Lastuvkova A, de Wit E, Klose RJ. Dimitrova E, et al. Nat Struct Mol Biol. 2022 Oct;29(10):1000-1010. doi: 10.1038/s41594-022-00840-5. Epub 2022 Oct 11. Nat Struct Mol Biol. 2022. PMID: 36220895 Free PMC article. - Indirect neurogenesis in space and time.
Thor S. Thor S. Nat Rev Neurosci. 2024 Aug;25(8):519-534. doi: 10.1038/s41583-024-00833-x. Epub 2024 Jul 1. Nat Rev Neurosci. 2024. PMID: 38951687 Review. - PHC3, a component of the hPRC-H complex, associates with E2F6 during G0 and is lost in osteosarcoma tumors.
Deshpande AM, Akunowicz JD, Reveles XT, Patel BB, Saria EA, Gorlick RG, Naylor SL, Leach RJ, Hansen MF. Deshpande AM, et al. Oncogene. 2007 Mar 15;26(12):1714-22. doi: 10.1038/sj.onc.1209988. Epub 2006 Sep 25. Oncogene. 2007. PMID: 17001316 Free PMC article. - Polycomb group complexes--many combinations, many functions.
Kerppola TK. Kerppola TK. Trends Cell Biol. 2009 Dec;19(12):692-704. doi: 10.1016/j.tcb.2009.10.001. Epub 2009 Nov 4. Trends Cell Biol. 2009. PMID: 19889541 Free PMC article. Review.
References
- Akasaka, T., M. Kanno, R. Balling, M. A. Mieza, M. Taniguchi, and H. Koseki. 1996. A role for mel-18, a Polycomb group-related vertebrate gene, during the anteroposterior specification of the axial skeleton. Development 122:1513-1522. - PubMed
- Akasaka, T., K. Tsuji, H. Kawahira, M. Kanno, K. Harigaya, L. Hu, Y. Ebihara, T. Nakahata, O. Tetsu, M. Taniguchi, and H. Koseki. 1997. The role of mel-18, a mammalian Polycomb group gene, during IL-7-dependent proliferation of lymphocyte precursors. Immunity 7:135-146. - PubMed
- Akasaka, T., M. van Lohuizen, N. van der Lugt, Y. Mizutani-Koseki, M. Kanno, M. Taniguchi, M. Vidal, M. Alkema, A. Berns, and H. Koseki. 2001. Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development 128:1587-1597. - PubMed
- Alkema, M. J., M. Bronk, E. Verhoeven, A. Otte, L. J. van 't Veer, A. Berns, and M. van Lohuizen. 1997. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian polycomb complex. Genes Dev. 11:226-240. - PubMed
- Atsuta, T., Y. Fujimura, H. Moriya, M. Vidal, T. Akasaka, and H. Koseki. 2001. Production of monoclonal antibodies against mammalian Ring1B proteins. Hybridoma 20:43-46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous