NFAT5 binds to the TNF promoter distinctly from NFATp, c, 3 and 4, and activates TNF transcription during hypertonic stress alone - PubMed (original) (raw)
NFAT5 binds to the TNF promoter distinctly from NFATp, c, 3 and 4, and activates TNF transcription during hypertonic stress alone
Jonathan H Esensten et al. Nucleic Acids Res. 2005.
Abstract
Tumor necrosis factor (TNF) is a pro-inflammatory cytokine that plays an important role in a variety of infectious and autoimmune disorders. Its transcription is regulated in a stimulus- and cell-type-specific manner via the recruitment of distinct DNA/activator complexes forming secondary structures or enhanceosomes. NFATp, a member of the nuclear factor of activated T cells (NFAT) family of transcription factors, plays a critical role in TNF gene regulation under a variety of conditions. In this study, we show that NFAT5, the most recently described NFAT family member, binds to the TNF promoter in a manner distinct from other NFAT proteins and is a key mediator in the activation of TNF gene transcription during hypertonic stress alone.
Figures
Figure 1
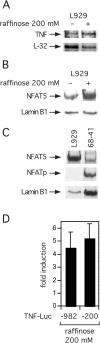
Hypertonicity induces TNF transcription and NFAT5 expression. (A) Induction of TNF mRNA by hypertonic stress. L929 cells were cultured in medium supplemented with 200 mM raffinose for 18 h. RNA was harvested and submitted to RNase protection assay. (B) Hypertonic stress up-regulates NFAT5 expression. L929 cells were cultured to ∼95% confluence in a 6-well plate and stimulated with complete medium or medium supplemented with 200 mM raffinose for 18 h, as indicated. Whole cell extracts were collected and submitted to western blot analysis. Results are representative of four independent experiments. (C) L929 cells do not express NFATp. Whole cell extract (30 μg) from 68 to 41 cells were submitted to western blot analysis as a positive control for NFATp expression. L929 cells were stimulated with hypertonic stress as above. Results are representative of two independent experiments. Lamin B1 was used as a loading control. (D) Induction of TNF luciferase activity by hypertonic stress in L929 fibroblasts. A total of 2 × 106 L929 cells were transfected with 1 μg −200 TNF-Luc or −982 TNF-Luc and 0.4 μg pTK-RL control vectors by DEAE-dextran. Cells were stimulated with complete medium supplemented with 200 mM raffinose 24 h after transfection. Cells were harvested 18 h after stimulation and luciferase activities were assayed. Results are from four independent experiments. Error bars are the standard deviation.
Figure 2
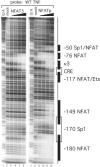
NFAT5 binds to the proximal TNF promoter in a pattern distinct from NFATp. DNase I footprinting assay of the TNF promoter with NFAT5. The DBA of recombinant human NFAT5 (DBD5) binds to two sites on the proximal TNF promoter between −120 and +87 nt relative to the transcription start site. Of the two sites, the −76 NFAT binding site binds at lower protein concentrations than the κ3 site. Increasing concentrations of DBD5 (25 ng, 125 ng, 500 ng and 2.5 μg) are represented by the wedge over lanes 3–6. Lighter bands indicate regions of reduced cleavage by DNase I due to protein binding. Several known transcription factor binding sites and their locations are indicated to the right of the gel. Recombinant NFATp binds six sites in the proximal promoter region (see lanes 9–12).
Figure 3
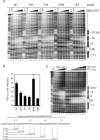
NFAT family members bind portions of the TNF promoter with different affinities. (A) EMSA using recombinant NFATp, NFATc, NFAT3 and NFAT4 with portions of the TNF promoter. Three probes (−76, κ3L and κ3LL) were end-labeled with 32P and incubated with recombinant protein, as indicated. Free probe marked at the bottom of the gel indicates equal loading. (B) EMSA using recombinant DBD5 or DBD5-GST and probes with portions of the TNF promoter. A diagram of the proximal human TNF promoter with probes for EMSA is just below. Known binding elements are boxed. Putative NFAT5 binding sites, based on the known consensus sequence, are bracketed. (C) EMSA using recombinant NFATp, NFATc, NFAT3, NFAT4 and NFAT5 with portions of the TNF promoter. Three probes (κ3L, κ3LL and −180) were end-labeled with 32P and incubated with recombinant protein, as indicated. Free probe marked at the bottom of the gel indicates equal loading. (D) EMSA using raffinose-stimulated L929 nuclear extracts and the κ3LL probe. Nuclear extracts were prepared from L929 cells unstimulated (−) or stimulated (+) with raffinose (200 mM) for 18 h. An oligonucleotide probe containing the −76 NFAT, κ3-NFAT and the CRE binding sites (κ3LL) was used and the binding assay was performed in the presence or absence of antibodies to NFAT5, NFATc, ATF-2, c-jun or isotype control (normal rabbit serum) as indicated in the figure. The complexes containing NFAT5 or ATF-2 alone are indicated by the gray arrows, and the composite NFAT5/ATF-2/c-jun complexes are indicated by the black arrows in the figure.
Figure 4
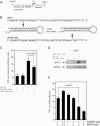
Mutations in the −76 NFAT and κ3 site, but not in the −84-Ets site, decrease binding of recombinant NFAT5 and hypertonic induction of TNF. (A) DNase I footprinting assay of mutated TNF promoter with NFAT5 (DBD5-GST). The 3′M and 5′M sites are in the κ3 element and the −76 mutation is in the −76 NFAT site. The −84 mutation is between these two sites. For a diagram, see the bottom of the figure. (B) Relative activity of TNF luciferase constructs containing mutations in known binding sites of transcriptional activators. A total of 5 × 106 L929 cells were transfected with 1 μg −200 TNF-Luc and 0.2 μg pTK-RL control vectors by DEAE-dextran. Cells were stimulated with complete medium supplemented with 200 mM raffinose 24 h after transfection. Cells were harvested 18 h after stimulation and Photinus luciferase and Renilla activities were assayed. (C) DNase I footprinting assay of competitive binding of NFAT5 and ATF-2/c-jun. The binding of ANF-2/c-jun to the CRE element was not blocked by increasing amounts of DBD5-GST (see lanes 13–17).
Figure 5
A vector expressing a shRNA targeting NFAT5 and a dominant negative NFAT5 down-regulate hypertonic induction of TNF luciferase activity. (A) Diagram of the shRNA-expressing construct. The murine U6 small nuclear RNA (snRNA) promoter was cloned into pBluescript followed by a sequence coding for the shRNA targeting murine NFAT5. See Materials and Methods for details. (B) Diagram of the shRNAs expressed from the scrambled sequence (U6-N) and the sequence targeting exon 8 of murine NFAT5 (U6-N5 ex8). (C and D) A vector expressing an shRNA against NFAT5 depresses hypertonic induction of TNF-Luc activity in L929 cells and lowers NFAT5 levels. A total of 3 × 105 cells were plated in 6-wells plates (∼95% confluent) and were transfected the next day with 0.7 μg TNF-Luc, 0.3 μg of TK-RL and 1 μg of either the U6-N or the U6-N5 ex8 vector. The cells were stimulated as indicated 36 h later with complete medium or medium supplemented with 200 mM raffinose for 18 h. Cells were harvested and assayed as above. Error bars are the SD. The _P_-value was calculated with an unpaired two-tailed _t_-test. Results are from four independent experiments. (D) Nuclear extracts from mock or raffinose treated L929 cells transfected with the U6 constructs were also analyzed by western blot analysis with an antibody to NFAT5, which demonstrates specific knockdown of NFAT5 protein levels by the U6-N5 ex8 vector as shown. (E) A vector expressing a dominant negative NFAT5 decreases hypertonic induction of TNF-Luc activity. L929 cells were co-transfected by DEAE-dextran with TNF-Luc, pTK-RL, and different amounts of either pcDNA3, N5-DN or both vectors as indicated. Cells were stimulated and extracts were assayed as above. The _P_-value was calculated with an unpaired two-tailed _t_-test. Results are from four independent experiments.
Similar articles
- Transcriptional control of the TNF gene.
Falvo JV, Tsytsykova AV, Goldfeld AE. Falvo JV, et al. Curr Dir Autoimmun. 2010;11:27-60. doi: 10.1159/000289196. Epub 2010 Feb 18. Curr Dir Autoimmun. 2010. PMID: 20173386 Free PMC article. Review. - Cell-type-specific regulation of the human tumor necrosis factor alpha gene in B cells and T cells by NFATp and ATF-2/JUN.
Tsai EY, Yie J, Thanos D, Goldfeld AE. Tsai EY, et al. Mol Cell Biol. 1996 Oct;16(10):5232-44. doi: 10.1128/MCB.16.10.5232. Mol Cell Biol. 1996. PMID: 8816436 Free PMC article. - Molecular mechanism of NFAT family proteins for differential regulation of the IL-2 and TNF-alpha promoters.
Oum JH, Han J, Myung H, Hleb M, Sharma S, Park J. Oum JH, et al. Mol Cells. 2002 Feb 28;13(1):77-84. Mol Cells. 2002. PMID: 11911478 - The role of NFATp in cyclosporin A-sensitive tumor necrosis factor-alpha gene transcription.
McCaffrey PG, Goldfeld AE, Rao A. McCaffrey PG, et al. J Biol Chem. 1994 Dec 2;269(48):30445-50. J Biol Chem. 1994. PMID: 7982959 - The role of NFAT5/TonEBP in establishing an optimal intracellular environment.
Ho SN. Ho SN. Arch Biochem Biophys. 2003 May 15;413(2):151-7. doi: 10.1016/s0003-9861(03)00130-9. Arch Biochem Biophys. 2003. PMID: 12729611 Review.
Cited by
- Transcriptional control of the TNF gene.
Falvo JV, Tsytsykova AV, Goldfeld AE. Falvo JV, et al. Curr Dir Autoimmun. 2010;11:27-60. doi: 10.1159/000289196. Epub 2010 Feb 18. Curr Dir Autoimmun. 2010. PMID: 20173386 Free PMC article. Review. - NFAT5 regulates HIV-1 in primary monocytes via a highly conserved long terminal repeat site.
Ranjbar S, Tsytsykova AV, Lee SK, Rajsbaum R, Falvo JV, Lieberman J, Shankar P, Goldfeld AE. Ranjbar S, et al. PLoS Pathog. 2006 Dec;2(12):e130. doi: 10.1371/journal.ppat.0020130. PLoS Pathog. 2006. PMID: 17173480 Free PMC article. - PPE38 of Mycobacterium marinum triggers the cross-talk of multiple pathways involved in the host response, as revealed by subcellular quantitative proteomics.
Wang H, Dong D, Tang S, Chen X, Gao Q. Wang H, et al. J Proteome Res. 2013 May 3;12(5):2055-66. doi: 10.1021/pr301017e. Epub 2013 Apr 8. J Proteome Res. 2013. PMID: 23514422 Free PMC article. - TonEBP regulates the hyperosmotic expression of aquaporin 1 and 5 in the intervertebral disc.
Snuggs JW, Tessier S, Bunning RAB, Shapiro IM, Risbud MV, Le Maitre CL. Snuggs JW, et al. Sci Rep. 2021 Feb 4;11(1):3164. doi: 10.1038/s41598-021-81838-9. Sci Rep. 2021. PMID: 33542263 Free PMC article. - TNF inhibits NKCC2 phosphorylation by a calcineurin-dependent pathway.
Hao S, Lasaracina AP, Epps J, Ferreri NR. Hao S, et al. Am J Physiol Renal Physiol. 2025 Apr 1;328(4):F489-F500. doi: 10.1152/ajprenal.00251.2024. Epub 2025 Mar 10. Am J Physiol Renal Physiol. 2025. PMID: 40062390 Free PMC article.
References
- Tsai E.Y., Falvo J.V., Tsytsykova A.V., Barczak A.K., Reimold A.M., Glimcher L.H., Fenton M.J., Gordon D.C., Dunn I.F., Goldfeld A.E. A lipopolysaccharide-specific enhancer complex involving Ets, Elk-1, Sp1, and CREB binding protein and p300 is recruited to the tumor necrosis factor alpha promoter in vivo. Mol. Cell. Biol. 2000;20:6084–6094. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM056492/GM/NIGMS NIH HHS/United States
- CA42471/CA/NCI NIH HHS/United States
- R01 CA042471/CA/NCI NIH HHS/United States
- R37 CA042471/CA/NCI NIH HHS/United States
- GM056492/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources