Genetic analysis of Pten and Tsc2 functional interactions in the mouse reveals asymmetrical haploinsufficiency in tumor suppression - PubMed (original) (raw)
Genetic analysis of Pten and Tsc2 functional interactions in the mouse reveals asymmetrical haploinsufficiency in tumor suppression
Li Ma et al. Genes Dev. 2005.
Abstract
The role of tumor suppressor haploinsufficiency in oncogenesis is still poorly understood. The PTEN and TSC2 tumor suppressors function to antagonize mTOR (mammalian target of rapamycin) activation by Akt; hence, compound heterozygous inactivation of Pten and Tsc2 in the mouse may in principle exacerbate the tumor phenotypes observed in the single mutants in a reciprocal manner. In contrast, we found that while Tsc2 heterozygosity unmasks Pten haploinsufficiency in growth and tumor suppression, tumorigenesis in Tsc2+/- mutants is surprisingly not accelerated by Pten heterozygosity, even though mTOR activation is cooperatively enhanced by compound Pten/Tsc2 heterozygosity. We show that the wild-type alleles of both Pten and Tsc2 are retained in prostate tumors from both Pten+/- and Pten+/-Tsc2+/- mice, whereas TSC-related tumor lesions are invariably associated with Tsc2 loss of heterozygosity (LOH) in both Tsc2+/- and Pten+/-Tsc2+/- mice. These findings demonstrate that inactivation of TSC2 is epistatic to PTEN in the control of tumor initiation and progression and, importantly, that both Pten and Tsc2 are haploinsufficient for suppression of tumorigenesis initiated by Pten heterozygosity, while neither Pten nor Tsc2 is haploinsufficient for repression of carcinogenesis arising from Tsc2 heterozygosity, providing a rationale for the differential cancer susceptibility of the two human conditions associated with PTEN or TSC2 heterozygous mutations.
Figures
Figure 1.
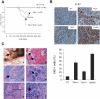
Reduction of the Tsc2 dose leads to reduction of survival, enhanced lymphoid proliferation, and development of skin cancer in Pten+/- mice. (A) Kaplan-Meier overall survival analysis shows a significant difference between Pten+/-Tsc2+/- and Pten+/- mice (P = 0.03). (B, upper panel) Ki-67 staining reveals increased proliferation in the lymph node upon compound loss of Pten and Tsc2 (200×). A representative count in triplicate, out of three with similar results, is shown in the lower panel with standard deviations. (C) Pathological features of a large skin lesion with squamous cell carcinoma in a 9-mo-old Pten+/-Tsc2+/- male mouse. (Panel a) The arrow points to an ulcerated skin lesion on the hind limb. (Panel b) Two detached squamous islands are indicated by the arrows (200×). (Panel c) A squamous pearl with central keratinization is shown (arrow; 200×). (Panel d) The arrow points at mitotic figures (600×). (Panel e) Disorganized basal cells with atypia and dyskeratosis (100×). (Panel f) A magnified image (400×) of the boxed region in panel e. The solid arrow indicates nuclear atypia, and the open arrow indicates dyskeratosis.
Figure 2.
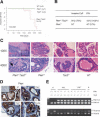
Reduction of the Tsc2 dose in Pten+/- mice leads to development of invasive CaP, which retains both Pten and Tsc2 wild-type alleles. (A) Kaplan-Meier disease-free survival curve of invasive CaP. (B) Incidence of invasive CaP and PIN in Pten+/-Tsc2+/- and Pten+/- mice. (C) Prostate tumorigenesis in AP of a 9-mo-old Pten+/-Tsc2+/- mouse and comparative analysis of littermates of various genotypes (100×, 400×). The boxed region and arrow indicate the area of invasion. (D) Expression of Pten in AP of 8-mo-old littermates (200×). Note the comparable level of Pten staining in neoplastic lesions (indicated by arrows) and in adjacent preneoplastic tissues. (Inset) Pten staining of a prostate from a Pten prostate conditional knockout mouse. (E) LOH analysis of Pten and Tsc2 in prostate tumors. Genomic DNA was extracted from laser capture-microdissected prostate tumors and normal counterpart, and amplified by PCR to detect wild-type (wt) and mutated (mut) alleles of Pten and Tsc2, respectively. (N) Normal prostate cells.
Figure 3.
Renal carcinogenesis in Pten+/-Tsc2+/- mice is associated with Tsc2 LOH and is not accelerated by reduction of the Pten dose. (A) Kaplan-Meier disease-free survival curve of renal cystadenoma. (B) Kaplan-Meier disease-free survival curve of renal adenocarcinoma. (C) Incidence of renal cystadenoma and renal carcinoma in Pten+/-Tsc2+/- and Tsc2+/- mice. (D) H&E sections show a papillary carcinoma in the kidney of a 5-mo-old Pten+/-Tsc2+/- mouse, and a papillary cystic carcinoma in the kidney of a Tsc2+/- littermate (40×, 400×). (E) LOH analysis of Pten and Tsc2 in renal carcinomas. Genomic DNA was extracted from laser capture-microdissected renal carcinomas and a normal counterpart, and amplified by PCR to detect wild-type (wt) and mutated (mut) alleles of Pten and Tsc2, respectively. (RC) Renal carcinoma; (N) normal kidney cells; (-) no template.
Figure 4.
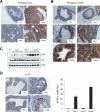
Biochemical and biological effects of compound loss of Pten and Tsc2 in the prostate. (A) Marked increase in phospho-Akt level in the prostates of either Pten+/- or Pten+/-Tsc2+/- mice (200×). (B) Phospho-mTOR level is markedly increased only in the prostates of Pten+/-Tsc2+/- mice (200×). A PIN lesion from a Pten+/- mouse and a prostate carcinoma from a Pten+/-Tsc2+/- mouse are shown along with normal epithelium from all genotypes. (C) Western blot analysis of prostate homogenates of various genotypes shows a marked increase in phospho-S6 level in the prostate of Pten+/-Tsc2+/- mice. (D, left panel) Ki-67 staining reveals increased proliferation in the prostate upon compound loss of Pten and Tsc2 (400×). Note that the area with prominent Ki-67 signals in the prostate of Pten+/-Tsc2+/- mice is preneoplastic, while the area shown in the prostate of Pten+/- mice is a PIN lesion. A representative count in triplicate out of three with similar results is shown in the right panel with standard deviations.
Figure 5.
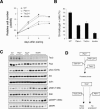
Biochemical and biological effects of compound loss of Pten and Tsc2 in MEFs. (A) Growth curves of MEFs at passage 3 show a growth advantage of Pten+/-Tsc2+/- and Pten+/- MEFs over either Tsc2+/- or wild-type MEFs. A representative experiment in triplicate out of three with similar results is shown with standard deviations. (B) SA-β-gal staining of MEFs at passage 10 shows bypass of cellular senescence in Pten+/-Tsc2+/- MEFs. A representative experiment in triplicate out of three with similar results is shown with standard deviations. (C) Western blot analysis of Tsc2, Pten, phospho-Akt, phospho-S6K, and phospho-4EBP1 levels in primary MEFs of various genotypes. (D) Model of asymmetrical haploinsufficiency and the nonreciprocal relationship between Pten and Tsc2 in tumor suppression. While tumorigenesis initiated by Pten heterozygous inactivation (e.g., PIN) undergoes progression to cancer upon either Pten LOH or partial loss of another tumor suppressor gene (e.g., Tsc2, or Cdkn1b encoding the p27kip1 protein), TSC-associated tumor lesions progress to full-blown malignancy only upon complete loss of the Tsc2 gene.
Similar articles
- Feedback inhibition of Akt signaling limits the growth of tumors lacking Tsc2.
Manning BD, Logsdon MN, Lipovsky AI, Abbott D, Kwiatkowski DJ, Cantley LC. Manning BD, et al. Genes Dev. 2005 Aug 1;19(15):1773-8. doi: 10.1101/gad.1314605. Epub 2005 Jul 18. Genes Dev. 2005. PMID: 16027169 Free PMC article. - Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis.
Kim MJ, Cardiff RD, Desai N, Banach-Petrosky WA, Parsons R, Shen MM, Abate-Shen C. Kim MJ, et al. Proc Natl Acad Sci U S A. 2002 Mar 5;99(5):2884-9. doi: 10.1073/pnas.042688999. Epub 2002 Feb 19. Proc Natl Acad Sci U S A. 2002. PMID: 11854455 Free PMC article. - Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis.
Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Pandolfi PP. Chen Z, et al. Nature. 2005 Aug 4;436(7051):725-30. doi: 10.1038/nature03918. Nature. 2005. PMID: 16079851 Free PMC article. - The role of PTEN in the progression and survival of prostate cancer.
Deocampo ND, Huang H, Tindall DJ. Deocampo ND, et al. Minerva Endocrinol. 2003 Jun;28(2):145-53. Minerva Endocrinol. 2003. PMID: 12717346 Review. - The biology and clinical relevance of the PTEN tumor suppressor pathway.
Sansal I, Sellers WR. Sansal I, et al. J Clin Oncol. 2004 Jul 15;22(14):2954-63. doi: 10.1200/JCO.2004.02.141. J Clin Oncol. 2004. PMID: 15254063 Review.
Cited by
- Identification of structural aberrations in cancer by SNP array analysis.
Heinrichs S, Look AT. Heinrichs S, et al. Genome Biol. 2007;8(7):219. doi: 10.1186/gb-2007-8-7-219. Genome Biol. 2007. PMID: 17666119 Free PMC article. Review. - Determinants of sensitivity and resistance to rapamycin-chemotherapy drug combinations in vivo.
Wendel HG, Malina A, Zhao Z, Zender L, Kogan SC, Cordon-Cardo C, Pelletier J, Lowe SW. Wendel HG, et al. Cancer Res. 2006 Aug 1;66(15):7639-46. doi: 10.1158/0008-5472.CAN-06-0419. Cancer Res. 2006. PMID: 16885364 Free PMC article. - Feedback inhibition of Akt signaling limits the growth of tumors lacking Tsc2.
Manning BD, Logsdon MN, Lipovsky AI, Abbott D, Kwiatkowski DJ, Cantley LC. Manning BD, et al. Genes Dev. 2005 Aug 1;19(15):1773-8. doi: 10.1101/gad.1314605. Epub 2005 Jul 18. Genes Dev. 2005. PMID: 16027169 Free PMC article. - Akt-dependent activation of mTORC1 complex involves phosphorylation of mTOR (mammalian target of rapamycin) by IκB kinase α (IKKα).
Dan HC, Ebbs A, Pasparakis M, Van Dyke T, Basseres DS, Baldwin AS. Dan HC, et al. J Biol Chem. 2014 Sep 5;289(36):25227-40. doi: 10.1074/jbc.M114.554881. Epub 2014 Jul 2. J Biol Chem. 2014. PMID: 24990947 Free PMC article. - The Neurodevelopmental Pathogenesis of Tuberous Sclerosis Complex (TSC).
Feliciano DM. Feliciano DM. Front Neuroanat. 2020 Jul 14;14:39. doi: 10.3389/fnana.2020.00039. eCollection 2020. Front Neuroanat. 2020. PMID: 32765227 Free PMC article. Review.
References
- Cairns P., Okami, K., Halachmi, S., Halachmi, N., Esteller, M., Herman, J.G., Jen, J., Isaacs, W.B., Bova, G.S., and Sidransky, D. 1997. Frequent inactivation of PTEN/MMAC1 in primary prostate cancer. Cancer Res. 57: 4997-5000. - PubMed
- Di Cristofano A. and Pandolfi, P.P. 2000. The multiple roles of PTEN in tumor suppression. Cell 100: 387-390. - PubMed
- Di Cristofano A., Pesce, B., Cordon-Cardo, C., and Pandolfi, P.P. 1998. Pten is essential for embryonic development and tumour suppression. Nat. Genet. 19: 348-355. - PubMed
- Di Cristofano A., Kotsi, P., Peng, Y.F., Cordon-Cardo, C., Elkon, K.B., and Pandolfi, P.P. 1999. Impaired Fas response and autoimmunity in Pten+/- mice. Science 285: 2122-2125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA082328/CA/NCI NIH HHS/United States
- R01 CA-82328/CA/NCI NIH HHS/United States
- P50 CA092629/CA/NCI NIH HHS/United States
- U01 CA-84292/CA/NCI NIH HHS/United States
- U01 CA084292/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous