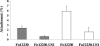Identification and characterization of a novel adhesin unique to oral fusobacteria - PubMed (original) (raw)
Identification and characterization of a novel adhesin unique to oral fusobacteria
Yiping W Han et al. J Bacteriol. 2005 Aug.
Abstract
Fusobacterium nucleatum is a gram-negative anaerobe that is prevalent in periodontal disease and infections of different parts of the body. The organism has remarkable adherence properties, binding to partners ranging from eukaryotic and prokaryotic cells to extracellular macromolecules. Understanding its adherence is important for understanding the pathogenesis of F. nucleatum. In this study, a novel adhesin, FadA (Fusobacterium adhesin A), was demonstrated to bind to the surface proteins of the oral mucosal KB cells. FadA is composed of 129 amino acid (aa) residues, including an 18-aa signal peptide, with calculated molecular masses of 13.6 kDa for the intact form and 12.6 kDa for the secreted form. It is highly conserved among F. nucleatum, Fusobacterium periodonticum, and Fusobacterium simiae, the three most closely related oral species, but is absent in the nonoral species, including Fusobacterium gonidiaformans, Fusobacterium mortiferum, Fusobacterium naviforme, Fusobacterium russii, and Fusobacterium ulcerans. In addition to FadA, F. nucleatum ATCC 25586 and ATCC 49256 also encode two paralogues, FN1529 and FNV2159, each sharing 31% identity with FadA. A double-crossover fadA deletion mutant, F. nucleatum 12230-US1, was constructed by utilizing a novel sonoporation procedure. The mutant had a slightly slower growth rate, yet its binding to KB and Chinese hamster ovarian cells was reduced by 70 to 80% compared to that of the wild type, indicating that FadA plays an important role in fusobacterial colonization in the host. Furthermore, due to its uniqueness to oral Fusobacterium species, fadA may be used as a marker to detect orally related fusobacteria. F. nucleatum isolated from other parts of the body may originate from the oral cavity.
Figures
FIG. 1.
I and II. Identification of F. nucleatum adhesins by far-Western analysis. a. F. nucleatum 12230 (I) or 40P (II) components stained with Coomassie blue following 12% SDS-PAGE. b. F. nucleatum 12230 (I) or 40P (II) components immobilized on PVDF membranes were incubated with streptavidin-HRP conjugate, followed by chemiluminescence reaction. c. F. nucleatum 12230 (I) or 40P (II) components immobilized on PVDF membranes were first incubated with biotinylated KB surface proteins, followed by incubation with streptavidin-HRP. “M,” protein size marker, with sizes indicated on the left; “NB,” nonboiled whole-cell F. nucleatum 12230 (I) or 40P (II); “B,” boiled whole-cell F. nucleatum 12230 (I) or 40P (II). The arrows point to bands visible in c but not in b, indicating binding to biotinylated KB proteins. The bottom arrow in Ic and the arrow in IIc indicate FadA. III. Competitive far-Western analysis. 40P or F. nucleatum 12230 whole-cell components immobilized on PVDF membranes were incubated directly with biotinylated KB surface proteins (a) or preincubated with 20× nonlabeled KB surface proteins prior to incubation with biotinylated KB surface proteins (b). “M,” protein size markers, as indicated on the left; lanes 1 to 5, 40P in increasing amounts (0.375, 0.625, 1.25, 2.5, and 5 μg, respectively); lanes 6, F. nucleatum 12230.
FIG. 2.
DNA dot blot analysis of the fadA gene in different Fusobacterium species. 1, F. gonidiaformans DUMC CF65-1; 2, F. gonidiaformans DUMC CF63-1; 3, F. mortiferum ATCC 25557; 4, F. naviforme DUMC CF108-1; 5, F. nucleatum ATCC 10953; 6, F. nucleatum ATCC 25586; 7, F. nucleatum ATCC 23726; 8, F. nucleatum 12230; 9, F. nucleatum ATCC 49256; 10, F. nucleatum ATCC 51190; 11, F. nucleatum PK1594; 12, F. nucleatum DUMC2929; 13, F. nucleatum DUMC3349; 14, F. nucleatum DUMC3156; 15, F. nucleatum DUMC1356; 16, F. nucleatum DUMC2079; 17, F. periodonticum ATCC 33693; 18, F. russii ATCC 25533; 19, F. simiae ATCC 33568; 20, F. ulcerans ATCC 49185.
FIG. 3.
Amino acid sequence alignment of FadA and its paralogues. Highlighted in gray are the identical residues shared among FadA proteins. The sequences of two FadA paralogues, FN1529 from F. nucleatum ATCC 25586 and FNV2159 from F. nucleatum ATCC 49256, are listed below FadA. The conserved and identical residues between FadA and the paralogues are indicated. Fn, F. nucleatum; Fp, F. periodonticum; Fs, F. simiae. The numbers above the sequence indicate amino acid positions in the secreted form of FadA, and the numbers beside the sequence indicate positions in the intact form.
FIG. 4.
Inactivation of the fadA gene of F. nucleatum 12230. A. Schematic diagram of construction of the Δ_fadA_::erm mutant by double-crossover allelic exchange. The erythromycin resistance cassette ermF-ermAM was inserted between bp 71 and 365 of the fadA gene. The shaded boxes represent regions hybridizing with the probe during Southern blotting. The sizes of the fragments hybridized with the probe are indicated. H, HindIII cleavage sites; E, EcoRI cleavage sites. B. Southern blot analysis of F. nucleatum 12230 and F. nucleatum 12230-US1, using a 359-bp fadA fragment as a probe. Lanes: 1, F. nucleatum 12230 digested with EcoRI; 2, F. nucleatum 12230-US1 digested with EcoRI; 3, F. nucleatum 12230 digested with HindIII; 4, F. nucleatum 12230-US1 digested with HindIII. The DNA size markers are indicated on the left. C. Western blot analysis of F. nucleatum 12230 and 12230-US1, using anti-FadA antibodies. Lanes: 1, protein size markers, with molecular masses shown on the left; 2, F. nucleatum 12230; 3, F. nucleatum 12230-US1. The arrow indicates FadA.
FIG. 5.
RT-PCR and Northern blot analyses of F. nucleatum 12230 and F. nucleatum 12230-US1. A. Schematic diagram showing locations of primers used for RT-PCR. The 2.4-kb _fadA_-containing fragment from F. nucleatum 12230 is presented as solid lines. The hairpin indicates the location of a putative transcription terminator. B. Northern blot analysis of F. nucleatum 12230 (lane 2) and F. nucleatum 12230-US1 (lane 3), using the 359-bp fadA fragment as a probe. Lane 1, 359-bp fadA fragment (positive control). C. RT-PCR analysis of expression of ORF2 (lanes 1 and 6), fadA (lanes 2 and 7), and ORF3 (lanes 3 and 8) in F. nucleatum 12230 (lanes 1 to 3) and F. nucleatum 12230-US1 (lanes 6 to 8). Lane 4, negative control without RNA; lane 5, 1.0-kb Plus DNA ladder.
FIG. 6.
Growth of F. nucleatum 12230 (solid triangles and solid line) and F. nucleatum 12230-US1 (open squares and dashed line) in Columbia broth. OD 600, optical density at 600 nm.
FIG. 7.
Attachment of F. nucleatum 12230 and F. nucleatum 12230-US1 to KB (hatched bars) and CHO (open bars) cells. The levels of attachment are means and standard deviations from three separate experiments, each performed in triplicate.
References
- Andersen, R. N., N. Ganeshkumar, and P. E. Kolenbrander. 1998. Helicobacter pylori adheres selectively to Fusobacterium spp. Oral Microbiol. Immunol. 13:51-54. - PubMed
- Babu, J. P., J. W. Dean, and M. J. Pabst. 1995. Attachment of Fusobacterium nucleatum to fibronectin immobilized on gingival epithelial cells or glass coverslips. J. Periodontol. 66:285-290. - PubMed
- Bauer, C., D. Schoonbroodt, C. Wagner, and Y. Horsmans. 2000. Liver abscesses due to Fusobacterium species. Liver 20:267-268. - PubMed
- Bekeredjian, R., S. Chen, P. A. Frenkel, P. A. Grayburn, and R. V. Shohet. 2003. Ultrasound-targeted microbubble destruction can repeatedly direct highly specific plasmid expression to the heart. Circulation 108:1022-1026. - PubMed
- Bolstad, A. I., B. T. Hogh, and H. B. Jensen. 1995. Molecular characterization of a 40-kDa outer membrane protein, FomA, of Fusobacterium periodonticum and comparison with Fusobacterium nucleatum. Oral Microbiol. Immunol. 10:257-264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DE 14447/DE/NIDCR NIH HHS/United States
- R03 DE014447/DE/NIDCR NIH HHS/United States
- R01 DE014924/DE/NIDCR NIH HHS/United States
- DE 09821/DE/NIDCR NIH HHS/United States
- R01 DE009821/DE/NIDCR NIH HHS/United States
- DE 14924/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases