Cross-talk between snurportin1 subdomains - PubMed (original) (raw)
Cross-talk between snurportin1 subdomains
Jason K Ospina et al. Mol Biol Cell. 2005 Oct.
Abstract
The initial steps of spliceosomal small nuclear ribonucleoprotein (snRNP) maturation take place in the cytoplasm. After formation of an Sm-core and a trimethylguanosine (TMG) cap, the RNPs are transported into the nucleus via the import adaptor snurportin1 (SPN) and the import receptor importin-beta. To better understand this process, we identified SPN residues that are required to mediate interactions with TMG caps, importin-beta, and the export receptor, exportin1 (Xpo1/Crm1). Mutation of a single arginine residue within the importin-beta binding domain (IBB) disrupted the interaction with importin-beta, but preserved the ability of SPN to bind Xpo1 or TMG caps. Nuclear transport assays showed that this IBB mutant is deficient for snRNP import but that import can be rescued by addition of purified survival of motor neurons (SMN) protein complexes. Conserved tryptophan residues outside of the IBB are required for TMG binding. However, SPN can be imported into the nucleus without cargo. Interestingly, SPN targets to Cajal bodies when U2 but not U1 snRNPs are imported as cargo. SPN also relocalizes to Cajal bodies upon treatment with leptomycin B. Finally, we uncovered an interaction between the N- and C-terminal domains of SPN, suggesting an autoregulatory function similar to that of importin-alpha.
Figures
Figure 1.
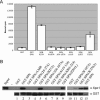
Schematic of SPN, alignment of SPN orthologues, and TMG cap binding assay. (A) Cartoon of SPN illustrating the TMG cap, Xpo1, and IBB. The IBB of SPN is defined as amino acid residues 26–65, based on similarity with the IBB of importin-α (Huber et al., 1998). The region of SPN responsible for Xpo1 binding activity has not been mapped and may not be a modular domain (Huber et al., 1998; Paraskeva et al., 1999). Based on proteolytic cleavage of SPN and UV cross-linking studies, the TMG-binding domain is thought to span residues 79–301 (Strasser et al., 2004). (B) Alignment of SPN orthologues. Human, frog, worm, fly, and plant SPN proteins are aligned, with identities shaded dark and similarities shaded light. A subset of the mutations used in this study is illustrated. Asterisks indicate alanine point substitutions and include R27, K32, R64, W107, and W276. Black bars indicate block alanine substitutions and include 25–27, 30–32, 43–45, 48–52, 63–64, 65–69, and 104–107. Gray bars indicate residues that were deleted and replaced with an amino acid linker consisting of IVAGS and include 39–52, 96–112, 119–134, 135–159, 170–187, 203–214, 255–262, and 266–279. The X indicates residue P291 that was mutated to leucine. Note that this alignment does not include the predicted C terminus of the Drosophila melanogaster orthologue. Carats (^) mark sites where one or more amino acids were excluded to facilitate the alignment. (C) Recombinant SPN can distinguish between m7G- and TMG-capped RNA. GST pulldowns were conducted using GST or GST-SPN and radiolabeled m7G- or TMG-capped U2 snRNA. After a 1-h incubation at 4°C, complexes were washed and bound counts determined.
Figure 2.
SPN mutants defective in TMG-cap binding also fail to interact with Xpo1. (A) Mutation of residue W107 or residues 104–107 abolish TMG binding. GST pulldowns were conducted using GST alone, GST-SPN, and the following GST-tagged SPN mutants: R27A, W107A, 104–107A, Δ119-134, Δ203-214, and P291L in the presence of radiolabeled, TMG-capped U2 snRNA. After incubation and washes, bound counts were determined using a scintillation counter. (B) SPN binding to Xpo1 is extremely sensitive to mutation. GST pulldowns were conducted using GST, GST-SPN (± LMB) and the following GST-tagged SPN mutants: 25-27A, R27A, 104-107A, W107A, Δ119-134, Δ203-214, W276A, and P291L in the presence of lysate expressing recombinant Xpo1-His and containing RanQ69L-GTP. Western blot analysis was conducted using anti-Xpo1 and anti-GST antibodies (loading control). Input shows 5% of the total lysate used in the pulldown.
Figure 3.
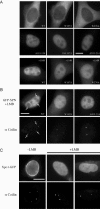
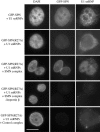
In vivo localization of SPN to Cajal bodies depends upon TMG binding but not Xpo1 binding. (A) SPN point mutants reduced for Xpo1 binding in vitro can interact with Xpo1 in vivo. The subcellular localization of GFP-SPN as well as mutant constructs W107A, and W276A were studied after transient transfection of HeLa cells in the presence or absence of LMB. GFP-tagged constructs bearing deletions or block substitutions (Δ119-134, 104-107A, and Δ203-214) were found to be nucleoplasmic in the absence of LMB treatment. (B) Xpo1 and TMG binding mutants fail to accumulate in Cajal bodies. HeLa cells were transiently transfected with wild-type GFP-SPN, -SPN(W107A), or -SPNΔ203-214 and treated with 20 nM LMB for 1 h. Immunofluorescence was then conducted with anti-coilin antibodies to localize Cajal bodies. Arrows mark Cajal bodies in the wild-type panel. (C) Xpo1 is enriched in Cajal bodies in the absence of LMB and is depleted from these nuclear bodies upon treatment. HeLa cells were transiently transfected with Xpo1-GFP and treated with 20 nM LMB for 1 h. Immunofluorescence was then conducted with anti-coilin antibodies to localize Cajal bodies. Bar, 10 μm.
Figure 4.
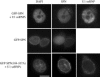
SPN import does not require bound cargo. The ability of GFP-SPN or -SPN(104-107A) to mediate U snRNP import was examined using an in vitro nuclear transport assay. Recombinant importin-β and purified Cy3-labeled U1 snRNPs were incubated with either wild-type or mutant His-GFP-SPN constructs and digitonin-permeabilized HeLa cells. Top, GFP-SPN and U1 snRNPs were efficiently imported. Bottom, GFP-SPN(104-107A) was imported into the nucleus (with pronounced enrichment at the nuclear rim). However, the mutant construct failed to mediate U1 import. Middle, GFP-SPN was imported in the absence of added U snRNPs, showing variable degrees of nuclear accumulation; some cells displayed prominent rim staining, whereas others were more uniformly labeled. Import reactions were incubated at 26°C for 35 min. Bar, 10 μm.
Figure 5.
SPN accumulates in Cajal bodies after import of U2 but not U1 snRNPs. The localizations of GFP-SPN and U snRNPs after snRNP import were determined using an in vitro nuclear transport assay. Recombinant importin-β and purified Cy3-labeled U1 or U2 snRNPs were incubated with wild-type His-GFP-SPN and digitonin-permeabilized HeLa cells. Cajal bodies were localized by immunofluorescence using an anti-coilin antibody. Bar, 10 μm.
Figure 6.
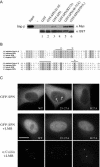
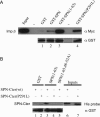
Mutants incapable of binding importin-β in vitro are efficiently imported in vivo. (A) Mutation of residue R27 disrupts SPN binding to importin-β. GST-pulldown assays were conducted using GST (negative control), GST-SPN, and the following SPN mutants: Δ1-65 (N-terminal deletion of 65 amino acids), R27A, 48-52A or P291L, along with recombinant His-myc-importin-β. Western analysis was conducted using anti-myc and anti-GST (loading control) antibodies. Input shows 10% of the total used in the pulldown. (B) Alignment of N-terminal regions of human importin-α and various SPN orthologues (human, Xenopus, and worm). Residues R27, K32, and R64 are marked with asterisks, regions 25–27, 30–32, 48–52, and 63–65 are overlined with bars. (C) Mutants deficient in importin-β binding are imported in vivo. HeLa cells were transiently transfected with wild-type GFP-SPN, or GFP-tagged SPN mutants 25-27A and R27A and treated with 20 nM LMB for 1 h. Immunofluorescence was then conducted with anti-coilin antibodies to localize Cajal bodies. Arrows indicate CBs in untreated cells. Bar, 10 μm.
Figure 7.
SPN binding to the TMG cap does not interfere with cap-independent import. Digitonin-permeabilized HeLa cells were incubated at 26°C for 35 min with purified Cy3-labeled U1 snRNPs, recombinant importin-β and either GFP-SPN or GFP-SPN(R27A) in the presence or absence of purified SMN complexes. Where indicated importin-β was omitted from the import reaction. Note that neither GFP-SPN(R27A) nor Cy3-U1 snRNPs were imported in the absence of added SMN complexes. Both were imported when 400 ng of purified SMN complexes were added along with T buffer and an ATP regenerating system (see Materials and Methods). Bar, 10 μm.
Figure 8.
SPN N- and C-terminal domains interact. (A) The binding of SPN to importin-β is increased upon mutation or truncation of the C terminus. Pulldown analysis was conducted using GST, GST-SPN, GST-SPN(1-65), or GST-SPN(P291L) and recombinant importin-β. The pulldowns were analyzed by Western blot and probed with anti-myc antibody or anti-GST as the loading control. Input shows 10% of the total used in the pulldown. (B) The N-terminal IBB domain and C terminus of SPN interact. GST pulldown assays were conducted using GST (negative control), GST-SPN(1-65), or GST-SPN(1-65) containing the 48-52 mutation, referred to as GST(1-65,48-52A). Lysates containing recombinant His-SPN(65-360), referred to as SPN-Cter(wt), or His-SPN(65-360) containing the P291L mutation, referred to as SPN-Cter(P291L) were used in the binding experiments. Western blot analysis was conducted and the blot probed with a His-probe or anti-GST antibody (loading control). Inputs show 1% of the total lysate used in the pulldown.
Similar articles
- SMN, the spinal muscular atrophy protein, forms a pre-import snRNP complex with snurportin1 and importin beta.
Narayanan U, Ospina JK, Frey MR, Hebert MD, Matera AG. Narayanan U, et al. Hum Mol Genet. 2002 Jul 15;11(15):1785-95. doi: 10.1093/hmg/11.15.1785. Hum Mol Genet. 2002. PMID: 12095920 Free PMC article. - Identification and characterization of Drosophila Snurportin reveals a role for the import receptor Moleskin/importin-7 in snRNP biogenesis.
Natalizio AH, Matera AG. Natalizio AH, et al. Mol Biol Cell. 2013 Sep;24(18):2932-42. doi: 10.1091/mbc.E13-03-0118. Epub 2013 Jul 24. Mol Biol Cell. 2013. PMID: 23885126 Free PMC article. - The importin-beta binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro.
Huber J, Dickmanns A, Lührmann R. Huber J, et al. J Cell Biol. 2002 Feb 4;156(3):467-79. doi: 10.1083/jcb.200108114. Epub 2002 Jan 28. J Cell Biol. 2002. PMID: 11815630 Free PMC article. - The importin β binding domain as a master regulator of nucleocytoplasmic transport.
Lott K, Cingolani G. Lott K, et al. Biochim Biophys Acta. 2011 Sep;1813(9):1578-92. doi: 10.1016/j.bbamcr.2010.10.012. Epub 2010 Oct 26. Biochim Biophys Acta. 2011. PMID: 21029753 Free PMC article. Review. - Dynamics of galectin-3 in the nucleus and cytoplasm.
Haudek KC, Spronk KJ, Voss PG, Patterson RJ, Wang JL, Arnoys EJ. Haudek KC, et al. Biochim Biophys Acta. 2010 Feb;1800(2):181-9. doi: 10.1016/j.bbagen.2009.07.005. Epub 2009 Jul 16. Biochim Biophys Acta. 2010. PMID: 19616076 Free PMC article. Review.
Cited by
- The Sm-core mediates the retention of partially-assembled spliceosomal snRNPs in Cajal bodies until their full maturation.
Roithová A, Klimešová K, Pánek J, Will CL, Lührmann R, Stanek D, Girard C. Roithová A, et al. Nucleic Acids Res. 2018 Apr 20;46(7):3774-3790. doi: 10.1093/nar/gky070. Nucleic Acids Res. 2018. PMID: 29415178 Free PMC article. - Biallelic variants in SNUPN cause a limb girdle muscular dystrophy with myofibrillar-like features.
Iruzubieta P, Damborenea A, Ioghen M, Bajew S, Fernandez-Torrón R, Töpf A, Herrero-Reiriz Á, Epure D, Vill K, Hernández-Laín A, Manterola M, Azkargorta M, Pikatza-Menoio O, Pérez-Fernandez L, García-Puga M, Gaina G, Bastian A, Streata I, Walter MC, Müller-Felber W, Thiele S, Moragón S, Bastida-Lertxundi N, López-Cortajarena A, Elortza F, Gereñu G, Alonso-Martin S, Straub V, de Sancho D, Teleanu R, López de Munain A, Blázquez L. Iruzubieta P, et al. Brain. 2024 Aug 1;147(8):2867-2883. doi: 10.1093/brain/awae046. Brain. 2024. PMID: 38366623 Free PMC article. - Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis.
Strzelecka M, Trowitzsch S, Weber G, Lührmann R, Oates AC, Neugebauer KM. Strzelecka M, et al. Nat Struct Mol Biol. 2010 Apr;17(4):403-9. doi: 10.1038/nsmb.1783. Epub 2010 Mar 28. Nat Struct Mol Biol. 2010. PMID: 20357773 - The traffic of proteins between nucleolar organizer regions and prenucleolar bodies governs the assembly of the nucleolus at exit of mitosis.
Muro E, Gébrane-Younís J, Jobart-Malfait A, Louvet E, Roussel P, Hernandez-Verdun D. Muro E, et al. Nucleus. 2010 Mar-Apr;1(2):202-11. doi: 10.4161/nucl.1.2.11334. Epub 2010 Jan 28. Nucleus. 2010. PMID: 21326952 Free PMC article. - Gemin4 is an essential gene in mice, and its overexpression in human cells causes relocalization of the SMN complex to the nucleoplasm.
Meier ID, Walker MP, Matera AG. Meier ID, et al. Biol Open. 2018 Feb 1;7(2):bio032409. doi: 10.1242/bio.032409. Biol Open. 2018. PMID: 29371219 Free PMC article.
References
- Adam, S. A., and Gerace, L. (1991). Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell 66, 837-847. - PubMed
- Andrade, M. A., Petosa, C., O'Donoghue, S. I., Muller, C. W., and Bork, P. (2001). Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309, 1-18. - PubMed
- Askjaer, P., Jensen, T. H., Nilsson, J., Englmeier, L., and Kjems, J. (1998). The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J. Biol. Chem. 273, 33414-33422. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM008613/GM/NIGMS NIH HHS/United States
- R01 GM053034/GM/NIGMS NIH HHS/United States
- S10 RR021228/RR/NCRR NIH HHS/United States
- R01 NS041617/NS/NINDS NIH HHS/United States
- T32-GM08613/GM/NIGMS NIH HHS/United States
- R01-GM53034/GM/NIGMS NIH HHS/United States
- S10 RR017980/RR/NCRR NIH HHS/United States
- R01-NS41617/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous