Structural basis of family-wide Rab GTPase recognition by rabenosyn-5 - PubMed (original) (raw)
Structural basis of family-wide Rab GTPase recognition by rabenosyn-5
Sudharshan Eathiraj et al. Nature. 2005.
Abstract
Rab GTPases regulate all stages of membrane trafficking, including vesicle budding, cargo sorting, transport, tethering and fusion. In the inactive (GDP-bound) conformation, accessory factors facilitate the targeting of Rab GTPases to intracellular compartments. After nucleotide exchange to the active (GTP-bound) conformation, Rab GTPases interact with functionally diverse effectors including lipid kinases, motor proteins and tethering complexes. How effectors distinguish between homologous Rab GTPases represents an unresolved problem with respect to the specificity of vesicular trafficking. Using a structural proteomic approach, we have determined the specificity and structural basis underlying the interaction of the multivalent effector rabenosyn-5 with the Rab family. The results demonstrate that even the structurally similar effector domains in rabenosyn-5 can achieve highly selective recognition of distinct subsets of Rab GTPases exclusively through interactions with the switch and interswitch regions. The observed specificity is determined at a family-wide level by structural diversity in the active conformation, which governs the spatial disposition of critical conserved recognition determinants, and by a small number of both positive and negative sequence determinants that allow further discrimination between Rab GTPases with similar switch conformations.
Conflict of interest statement
Competing interests statement. The authors declare that they have no competing financial interests.
Figures
Figure 1
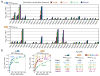
Mapping of the Rabenosyn-5 Rab GTPase binding domains. Mean Kd values and standard deviations were obtained from 2–4 SPR experiments using purified Rab GTPases and Rabenosyn-5 constructs. The oligomeric state (n = 1 for monomer; n = 2 for dimer) was determined from sedimentation equilibrium experiments over the concentration range from 5 – 50 mM. Values represent the mean and standard deviation for 3 measurements.
Figure 2
Quantitative family-wide analysis of Rab GTPase-effector specificity. a, Initial screen for the interaction of 6xHis (or GST) fusions of Rab GTPases with GST (or 6xHis) fusions of Rbsn440–503 and Rbsn728–784. For each potential interaction, the equilibrium SPR signal (Req) was measured at four concentrations of the 6xHis Rab GTPase or 6xHis Rbsn construct. b, Concentration dependence of the equilibrium SPR signal (Req) for the binding of 6xHis Rab GTPases to GST fusions of Rbsn440–503 and Rbsn728–784. Mean Kd values and standard deviations for 2–4 independent experiments are tabulated on the right.
Figure 3
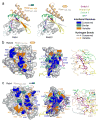
Structural basis of Rab recognition by Rabeonsoyn-5. a, Ribbon rendering of GTP-bound Rab4 Q67L and Rab22 Q64L in complex with the minimal Rab binding domains of Rabenosyn-5. b, Conservation and variability in the Rab22-Rbsn728–784 interface. Spheres covered by a semitransparent surface represent Rab22 (left panel) or Rbsn728–784 (right panel). Hydrogen bonding interactions are depicted in the right panel. c, Conservation and variability in the Rab4-Rbsn440–503 interface. Spheres covered by a semitransparent surface represent Rab4 (left panel) or Rbsn440–503 (right panel). Hydrogen bonding interactions are depicted in the right panel.
Figure 4
Structure-based mutational analysis of Rab and Rabeonsoyn-5 interaction specificity. a, Local alignment of representative Rab GTPases and Rabenosyn-5 homologs. Residues within the binding interfaces for Rab22-Rbsn728–784, Rab4-Rbsn440–503, Rab5-Rabaptin-5, and Rab3-Rabphilin are highlighted according to conservation. b, Effect of site specific substitutions on the interaction between Rab5 and Rbsn728–784 or Rab4 and Rbsn440–503. Mean values and standard deviations were calculated from 2 experiments. c, Views of the Rab22-Rbsn728–784 and Rab4-Rbsn440–503 interfaces relevant to the mutations discussed in the text. Also shown (middle panel) is the structure of the Rab5 G55Q mutant following superposition with Rab22.
Similar articles
- Structural basis for Rab GTPase recognition and endosome tethering by the C2H2 zinc finger of Early Endosomal Autoantigen 1 (EEA1).
Mishra A, Eathiraj S, Corvera S, Lambright DG. Mishra A, et al. Proc Natl Acad Sci U S A. 2010 Jun 15;107(24):10866-71. doi: 10.1073/pnas.1000843107. Epub 2010 Jun 1. Proc Natl Acad Sci U S A. 2010. PMID: 20534488 Free PMC article. - TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism.
Pan X, Eathiraj S, Munson M, Lambright DG. Pan X, et al. Nature. 2006 Jul 20;442(7100):303-6. doi: 10.1038/nature04847. Nature. 2006. PMID: 16855591 - Determinants of the broad recognition of exocytic Rab GTPases by Mss4.
Zhu Z, Delprato A, Merithew E, Lambright DG. Zhu Z, et al. Biochemistry. 2001 Dec 25;40(51):15699-706. doi: 10.1021/bi0116792. Biochemistry. 2001. PMID: 11747446 - Membrane recruitment of effector proteins by Arf and Rab GTPases.
Kawasaki M, Nakayama K, Wakatsuki S. Kawasaki M, et al. Curr Opin Struct Biol. 2005 Dec;15(6):681-9. doi: 10.1016/j.sbi.2005.10.015. Epub 2005 Nov 9. Curr Opin Struct Biol. 2005. PMID: 16289847 Review. - Diversity in structure and function of tethering complexes: evidence for different mechanisms in vesicular transport regulation.
Kümmel D, Heinemann U. Kümmel D, et al. Curr Protein Pept Sci. 2008 Apr;9(2):197-209. doi: 10.2174/138920308783955252. Curr Protein Pept Sci. 2008. PMID: 18393888 Review.
Cited by
- Unconventional endosome-like compartment and retromer complex in Toxoplasma gondii govern parasite integrity and host infection.
Sangaré LO, Alayi TD, Westermann B, Hovasse A, Sindikubwabo F, Callebaut I, Werkmeister E, Lafont F, Slomianny C, Hakimi MA, Van Dorsselaer A, Schaeffer-Reiss C, Tomavo S. Sangaré LO, et al. Nat Commun. 2016 Apr 11;7:11191. doi: 10.1038/ncomms11191. Nat Commun. 2016. PMID: 27064065 Free PMC article. - Human Golgi phosphoprotein 3 is an effector of RAB1A and RAB1B.
Cavieres VA, Cerda-Troncoso C, Rivera-Dictter A, Castro RI, Luchsinger C, Santibañez N, Burgos PV, Mardones GA. Cavieres VA, et al. PLoS One. 2020 Aug 13;15(8):e0237514. doi: 10.1371/journal.pone.0237514. eCollection 2020. PLoS One. 2020. PMID: 32790781 Free PMC article. - Structures of Drosophila melanogaster Rab2 and Rab3 bound to GMPPNP.
Lardong JA, Driller JH, Depner H, Weise C, Petzoldt A, Wahl MC, Sigrist SJ, Loll B. Lardong JA, et al. Acta Crystallogr F Struct Biol Commun. 2015 Jan 1;71(Pt 1):34-40. doi: 10.1107/S2053230X1402617X. Epub 2015 Jan 1. Acta Crystallogr F Struct Biol Commun. 2015. PMID: 25615965 Free PMC article. - Structural and Biophysical Characterization of Rab5a from Leishmania Donovani.
Maheshwari D, Yadav R, Rastogi R, Jain A, Tripathi S, Mukhopadhyay A, Arora A. Maheshwari D, et al. Biophys J. 2018 Oct 2;115(7):1217-1230. doi: 10.1016/j.bpj.2018.08.032. Epub 2018 Aug 30. Biophys J. 2018. PMID: 30241678 Free PMC article. - The glycine brace: a component of Rab, Rho, and Ran GTPases associated with hinge regions of guanine- and phosphate-binding loops.
Neuwald AF. Neuwald AF. BMC Struct Biol. 2009 Mar 5;9:11. doi: 10.1186/1472-6807-9-11. BMC Struct Biol. 2009. PMID: 19265520 Free PMC article.
References
- Pfeffer SR. Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 2001;11:487–491. - PubMed
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. - PubMed
- Sivars U, Aivazian D, Pfeffer SR. Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature. 2003;425:856–859. - PubMed
- Rak A, et al. Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science. 2003;302:646–650. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases