Formation of morphologically similar globular aggregates from diverse aggregation-prone proteins in mammalian cells - PubMed (original) (raw)
Formation of morphologically similar globular aggregates from diverse aggregation-prone proteins in mammalian cells
Hideyuki Mukai et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Huntington's disease is a progressive neurodegenerative disorder caused by a polyglutamine repeat expansion in the first exon of the huntingtin (Htt) protein. N-terminal Htt peptides with polyglutamine tracts in the pathological range (51-122 glutamines) form high-molecular-weight protein aggregates with fibrillar morphology in vitro, and they form discrete inclusion bodies in a cell-culture model. However, in some studies, formation of discrete Htt inclusions does not correlate well with cell death. We coexpressed N-terminal Htt fragments containing 91 glutamines fused to different affinity tags in HEK293 cells, and we isolated small aggregates by double sequential-affinity chromatography to assure the isolation of multimeric molecules. Transmission electron microscopy and atomic force microscopy revealed the isolated aggregates as globules or clusters of globules 4-50 nm in diameter without any detectable fibrillar species. Because small nonfibrillar oligomers, not mature fibrils, recently have been suggested to be the principal cytotoxic species in neurodegenerative disease, these Htt globular aggregates formed in cells may represent the pathogenic form of mutant Htt.
Figures
Fig. 1.
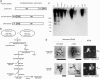
Isolation of HDQ91 aggregates under nondenaturing conditions. (a) Constructs. The GFP tag is indicated by a black box, and affinity tags (protein A and CBP) for isolation of HDQ91 aggregates are indicated by gray boxes. Q91, Htt exon 1 (3) protein with 91 glutamine repeats; TEV, TEV protease recognition sequence. (b) Diagram showing the isolation procedure for HDQ91 aggregates under nondenaturing conditions. (c) Immunoblotting (IB) of column fractions was performed using anti-GFP antibody (JL-8). Molecular mass markers are indicated in kDa at the left. The arrowhead indicates the bottom of the loading wells. (d) Silver staining of column fractions. The same amount of each indicated column fraction from c was applied to SDS/PAGE, and silver stained. (e) Fluorescence microscopy of column fractions. The individual fractions are as indicated in b. (Scale bar, 10 μm.)
Fig. 2.
Specificity of protein aggregation isolated by double-affinity method. The column fractions from a mixture of cells transfected with pQ91/GFP and pQ91/ProtA and cells transfected with only pQ91/CBP was immunoblotted by using anti-GFP antibody (JL-8). Molecular mass markers (in kDa) are indicated on the left. The naming of the individual fractions is the same as in Fig. 1_b_. The arrowhead indicates the bottom of loading wells.
Fig. 3.
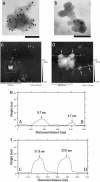
Structure of isolated HDQ91 aggregates. (a and b) TEM of uranyl acetate-stained immunogold-labeled HDQ91 aggregates. The size of the gold particles was 10 nm. (Scale bar, 100 nm.) (c) AFM height image of HDQ91 aggregates. (d) Magnified image of the boxed area in c. (e and f) The AFM surface profile along A–B and C–D axes in d, respectively. The arrows in d indicate the globular aggregates, corresponding to the arrows in e and f.
Fig. 4.
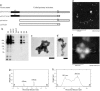
Isolation of HDQ91 aggregates in the presence of urea. (a) Constructs. Affinity tags [(His)6 tag and S·tag] for isolation of HDQ91 aggregates are indicated by gray boxes. Q91, Htt exon 1 (11) protein with 91 glutamine repeats; myc, myc tag; HA, HA tag; FLAG, FLAG tag. (b) Diagram showing the isolation step of HDQ91 aggregates in the presence of urea. (c) Immunoblotting of column fractions was performed by using anti-FLAG (M2) antibody. The individual fractions are as indicated in b. Molecular mass markers (in kDa) are indicated on the left. (d) TEM and AFM of HDQ91 aggregates isolated in the presence of urea. The size of the gold particles was 10 nm. (Scale bar, 100 nm.) The composition of the aggregates is indicated on the left.
Fig. 5.
Isolation of CFTRΔF508 aggregates under nondenaturing conditions. (a) Constructs. GFP tag is indicated by black box, and affinity tags (protein A and CBP) for isolation of CFTRΔF508 aggregates are indicated by gray boxes. TEV, TEV protease recognition sequence. (b) Immunoblotting of column fractions was performed by using anti-GFP antibody (JL-8). The individual fractions are designated by the convention introduced in Fig. 1_b_. Molecular mass markers (in kDa) are indicated on the left. (c and d) TEM of uranyl acetate-stained immunogold-labeled CFTRΔF508 aggregates. The size of the gold particles was 10 nm. (Scale bar, 100 nm.) (e–h) The AFM height images of CFTRΔF508 aggregates (e and f) and the surface profiles (g and h) along the A–B (e and g) and C–D (f and h) axes, respectively. The arrows in e indicate the globular aggregates, corresponding to the arrows in g. The arrows in f indicate the globular aggregates, corresponding to the arrows in h.
Similar articles
- Mutant huntingtin promotes the fibrillogenesis of wild-type huntingtin: a potential mechanism for loss of huntingtin function in Huntington's disease.
Busch A, Engemann S, Lurz R, Okazawa H, Lehrach H, Wanker EE. Busch A, et al. J Biol Chem. 2003 Oct 17;278(42):41452-61. doi: 10.1074/jbc.M303354200. Epub 2003 Jul 29. J Biol Chem. 2003. PMID: 12888569 - Huntingtin spheroids and protofibrils as precursors in polyglutamine fibrilization.
Poirier MA, Li H, Macosko J, Cai S, Amzel M, Ross CA. Poirier MA, et al. J Biol Chem. 2002 Oct 25;277(43):41032-7. doi: 10.1074/jbc.M205809200. Epub 2002 Aug 8. J Biol Chem. 2002. PMID: 12171927 - Atomic force microscopy analysis of the Huntington protein nanofibril formation.
Dahlgren PR, Karymov MA, Bankston J, Holden T, Thumfort P, Ingram VM, Lyubchenko YL. Dahlgren PR, et al. Nanomedicine. 2005 Mar;1(1):52-7. doi: 10.1016/j.nano.2004.11.004. Nanomedicine. 2005. PMID: 17292058 - Assessing mutant huntingtin fragment and polyglutamine aggregation by atomic force microscopy.
Burke KA, Godbey J, Legleiter J. Burke KA, et al. Methods. 2011 Mar;53(3):275-84. doi: 10.1016/j.ymeth.2010.12.028. Epub 2010 Dec 25. Methods. 2011. PMID: 21187152 - Mutant huntingtin fragments form oligomers in a polyglutamine length-dependent manner in vitro and in vivo.
Legleiter J, Mitchell E, Lotz GP, Sapp E, Ng C, DiFiglia M, Thompson LM, Muchowski PJ. Legleiter J, et al. J Biol Chem. 2010 May 7;285(19):14777-90. doi: 10.1074/jbc.M109.093708. Epub 2010 Mar 10. J Biol Chem. 2010. PMID: 20220138 Free PMC article.
Cited by
- Expression of the type VI intermediate filament proteins CP49 and filensin in the mouse lens epithelium.
FitzGerald P, Sun N, Shibata B, Hess JF. FitzGerald P, et al. Mol Vis. 2016 Aug 6;22:970-89. eCollection 2016. Mol Vis. 2016. PMID: 27559293 Free PMC article. - Prions Ex Vivo: What Cell Culture Models Tell Us about Infectious Proteins.
Krauss S, Vorberg I. Krauss S, et al. Int J Cell Biol. 2013;2013:704546. doi: 10.1155/2013/704546. Epub 2013 Oct 26. Int J Cell Biol. 2013. PMID: 24282413 Free PMC article. Review. - Protein quality control during erythropoiesis and hemoglobin synthesis.
Khandros E, Weiss MJ. Khandros E, et al. Hematol Oncol Clin North Am. 2010 Dec;24(6):1071-88. doi: 10.1016/j.hoc.2010.08.013. Hematol Oncol Clin North Am. 2010. PMID: 21075281 Free PMC article. Review. - Atomistic simulations of the effects of polyglutamine chain length and solvent quality on conformational equilibria and spontaneous homodimerization.
Vitalis A, Wang X, Pappu RV. Vitalis A, et al. J Mol Biol. 2008 Dec 5;384(1):279-97. doi: 10.1016/j.jmb.2008.09.026. Epub 2008 Sep 18. J Mol Biol. 2008. PMID: 18824003 Free PMC article. - The interplay between PolyQ and protein context delays aggregation by forming a reservoir of protofibrils.
Bulone D, Masino L, Thomas DJ, San Biagio PL, Pastore A. Bulone D, et al. PLoS One. 2006 Dec 27;1(1):e111. doi: 10.1371/journal.pone.0000111. PLoS One. 2006. PMID: 17205115 Free PMC article.
References
- Harper, P. (1996) Huntington's Disease (Saunders, Philadelphia).
- Vonsattel, J. P., Myers, R. H., Stevens, T. J., Ferrante, R. J., Bird, E. D. & Richardson, E. P., Jr., (1985) J. Neuropathol. Exp. Neurol. 44, 559-577. - PubMed
- The Huntington's Disease Collaborative Research Group (1993) Cell 72, 971-983. - PubMed
- Cooper, J. K., Schilling, G., Peters, M. F., Herring, W. J., Sharp, A. H., Kaminsky, Z., Masone, J., Khan, F. A., Delanoy, M., Borchelt, D. R., et al. (1998) Hum. Mol. Genet. 7, 783-790. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources