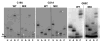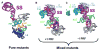Structural and functional analysis of 5S rRNA in Saccharomyces cerevisiae - PubMed (original) (raw)
Comparative Study
Structural and functional analysis of 5S rRNA in Saccharomyces cerevisiae
Sergey Kiparisov et al. Mol Genet Genomics. 2005 Oct.
Abstract
5S rRNA extends from the central protuberance of the large ribosomal subunit, through the A-site finger, and down to the GTPase-associated center. Here, we present a structure-function analysis of seven 5S rRNA alleles which are sufficient for viability in the yeast Saccharomyces cerevisiae when expressed in the absence of wild-type 5S rRNAs, and extend this analysis using a large bank of mutant alleles that show semi-dominant phenotypes in the presence of wild-type 5S rRNA. This analysis supports the hypothesis that 5S rRNA serves to link together several different functional centers of the ribosome. Data are also presented which suggest that in eukaryotic genomes selection has favored the maintenance of multiple alleles of 5S rRNA, and that these may provide cells with a mechanism to post-transcriptionally regulate gene expression.
Figures
Fig. 1
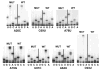
Direct rRNA sequence analyses of “pure” 5S rRNA mutants. rRNAs were extracted from ribosomes purified from JD1253 cells expressing wild-type or mutant forms of 5S rRNA. Oligonucleotides complementary to 5S rRNA were labeled with γ[32P]ATP using T4 polynucleotide kinase, and annealed with 5S rRNA isolated from purified ribosomes. Primer-extension reactions were performed using AMV reverse transcriptase, fractionated by electrophoresis through denaturing 12% polyacrylamide-urea gels, and labeled bands were visualized by autoradiography
Fig. 2
Growth phenotypes of “pure” 5S rRNA mutants. Mid-logarithmically growing JD1253 cells were diluted to 2 × 107 colony forming units (CFU)/ml. Subsequently, aliquots (5 μl) containing 104 CFU, and tenfold dilutions from the same cultures thereof were spotted either onto rich medium (YPAD) and incubated at 15, 30, and 37°C, or onto YPAD containing anisomycin (10 μg/ml) or sparsomycin (30 μg/ml) and incubated at 30°C
Fig. 3
a–d Effects of pure 5S rRNA mutants on programmed ribosomal frameshifting and virus propagation. JD1253 cells expressing the indicated “pure” 5S rRNA were assayed with respect to the following phenotypes. a Cells were transformed with 0-frame control and L–A derived −1 PRF test dual luciferase reporter plasmids, and −1 PRF efficiencies were determined as described by Harger and Dinman (2003). All assays were replicated at least 30 times, and standard errors (represented by the error bars) were calculated as described by Jacobs and Dinman (2004). b Programmed +1 ribosomal frameshifting was analyzed using a Ty_1_ derived +1 PRF signal cloned into the dual luciferase reporter plasmids as described above. c Total nucleic acids were extracted from JD1253 cells expressing the indicated 5S rRNA alleles, fractionated by electrophoresis on a 1.5% native agarose gel, and stained with ethidium bromide. Genomic DNA (gDNA), viral dsRNAs, and rRNAs are indicated. L-BC is a dsRNA virus unrelated to L–A and M1. d Cells were transformed with pJEF1105, a galactose-inducible Ty_1_ cDNA clone containing the neo_r_ selectable reporter, incubated at 20°C for 4 days on medium containing 2% galactose, replica plated onto medium containing 100 μg/ml 5-FOA, and incubated at 30°C to select for cells that had lost pJDF1105. Viable colonies were grown on YPAD medium overnight, and 10-fold dilutions of cells ranging from 106 to 102 CFU were spotted onto YPAD medium containing 100 μg/ml of Genticin. In parallel, 105–101 CFU were spotted onto YPAD medium alone
Fig. 4
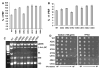
Chemical protection analyses. Ribosomes purified from JD1253 cells expressing either wild-type or mutant 5S rRNAs were chemically probed with DMS, kethoxal and CMCT. The products of reverse transcriptase primer extension reactions were subjected to PAGE analysis on denaturing PA-urea gels, and visualized by autoradiography. Left panel Representative autoradiographs showing altered reactivities of bases in 5S rRNA (top and middle), and 25S rRNA (bottom). Right panel Summary of the effects of pure mutants mapped onto 5S rRNA and the helix 95-sarcin/ricin loop (SRL) region of 25S rRNA. The open circles indicate positions of wild-type bases, and the arrows indicate the relevant mutant alleles. The color-coded bars indicate which mutant 5S rRNAs alter the chemical reactivities of other similarly coded, circled, bases in 5S and 25S rRNAs
Fig. 5
Direct rRNA sequence analyses of “mixed” 5S rRNA mutants. rRNAs were extracted from ribosomes purified from JD1111 cells expressing wild-type or mixtures of mutant and wild-type forms of 5S rRNA. Oligonucleotides complementary to 5S rRNA were labeled with γ[32P]ATP using T4 polynucleotide kinase, and annealed with 5S rRNA isolated from purified ribosomes. Primer-extension reactions were performed using AMV reverse transcriptase, fractionated by electrophoresis through denaturing 12% polyacrylamide-urea gels, and visualized by autoradiography. The asterisks indicate the 5S rRNA mutations in the mixed populations
Fig. 6
a, b Semi dominant effects of 5S rRNA alleles on programmed ribosomal frameshifting and virus propagation. a Effects on −1 PRF and killer virus propagation. b Effects on +1 PRF and Ty_1_ retrotransposition. JD1111 cells expressing wild-type or mixtures of wild-type and mutant 5S rRNA alleles were transformed with monocistronic lacZ_-based 0-frame, −1 or +1 frameshift reporter plasmids and L–A virus promoted −1, or Ty_1 promoted +1 PRF efficiencies were determined as previously described (Dinman et al. 1991; Dinman and Wickner 1992). Alleles of 5S rRNA are shown on the _X_-axis and PRF efficiencies are indicated on the Y_-axis. The error bars correspond to standard error. In panel a, the different shadings indicate inhibitory effects of 5S rRNA alleles on maintenance of either the M1 satellite virus alone or both L– A and M1, as previously described (Smith et al. 2001). In panel b, the shadings correspond to inhibitory effects of 5S rRNA alleles on Ty_1 retrotransposition frequencies assessed using pJEF1105
Fig. 7
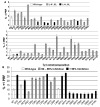
a, b Allelic variants of 5S rRNAs in eukaryotic genomes. a Conservation of multiple 5S rDNA alleles in eukaryotic genomes. GenBank was first queried for 5S rRNA sequences according to species, and the resulting sequences were used in BLAST searches (Altschul et al. 1990) to identify homologous sequences in the database. Sequences were hand curated to ensure their validity, and then aligned with one another as described in Materials and methods. RDN5-som and RDN5-ooc show the sequences of the hybrid yeast/Xenopus clones. Color coding is used to denote base substitutions. b Semi-dominant effects of naturally occurring yeast and hybrid yeast/Xenopus 5S rRNA alleles on L–A directed −1, and Ty_1_ promoted +1 PRF. Using the yeast RDN5-1 allele cloned into a high-copy-number 2μ vector, oligonucleotide-primed site-directed mutagenesis was used to create the other six naturally occurring yeast RDN5 alleles (RDN5-2 to RDN5-7), and the hybrid yeast/Xenopus RDN5-ooc and RDN5-som alleles (see Fig. 6a). These alleles were episomally expressed in JD932 cells, a wild-type yeast strain containing a full complement of chromosomal rDNA genes. Programmed −1 and +1 ribosomal frameshifting efficiencies were monitored as described in Fig. 6
Fig. 7
a, b Allelic variants of 5S rRNAs in eukaryotic genomes. a Conservation of multiple 5S rDNA alleles in eukaryotic genomes. GenBank was first queried for 5S rRNA sequences according to species, and the resulting sequences were used in BLAST searches (Altschul et al. 1990) to identify homologous sequences in the database. Sequences were hand curated to ensure their validity, and then aligned with one another as described in Materials and methods. RDN5-som and RDN5-ooc show the sequences of the hybrid yeast/Xenopus clones. Color coding is used to denote base substitutions. b Semi-dominant effects of naturally occurring yeast and hybrid yeast/Xenopus 5S rRNA alleles on L–A directed −1, and Ty_1_ promoted +1 PRF. Using the yeast RDN5-1 allele cloned into a high-copy-number 2μ vector, oligonucleotide-primed site-directed mutagenesis was used to create the other six naturally occurring yeast RDN5 alleles (RDN5-2 to RDN5-7), and the hybrid yeast/Xenopus RDN5-ooc and RDN5-som alleles (see Fig. 6a). These alleles were episomally expressed in JD932 cells, a wild-type yeast strain containing a full complement of chromosomal rDNA genes. Programmed −1 and +1 ribosomal frameshifting efficiencies were monitored as described in Fig. 6
Fig. 8
a–c Correlation of 5S rRNA structure with function. Mutations in 5S rRNA that produced phenotypic and physical changes are mapped onto the high-resolution structure of the yeast ribosome described by Spahn et al. (2004). a Mapping of sites affected in “pure” mutants onto 5S and 25S rRNA structures. Positions marked in green in 5S rRNA show locations of A20, A79, and U81. Positions marked in red show the locations of bases whose chemical reactivities were altered as a consequence of the 5S rRNA mutations. b, c “Mixed” 5S rRNA alleles that affected +1 (b) or −1 PRF (c). Mutations at bases shown in red stimulated PRF, and those shown in blue inhibited PRF
Similar articles
- Saturation mutagenesis of 5S rRNA in Saccharomyces cerevisiae.
Smith MW, Meskauskas A, Wang P, Sergiev PV, Dinman JD. Smith MW, et al. Mol Cell Biol. 2001 Dec;21(24):8264-75. doi: 10.1128/MCB.21.24.8264-8275.2001. Mol Cell Biol. 2001. PMID: 11713264 Free PMC article. - Structure and function of 5S rRNA in the ribosome.
Bogdanov AA, Dontsova OA, Dokudovskaya SS, Lavrik IN. Bogdanov AA, et al. Biochem Cell Biol. 1995 Nov-Dec;73(11-12):869-76. doi: 10.1139/o95-094. Biochem Cell Biol. 1995. PMID: 8722002 Review. - Identification of the gene encoding the 5S ribosomal RNA maturase in Bacillus subtilis: mature 5S rRNA is dispensable for ribosome function.
Condon C, Brechemier-Baey D, Beltchev B, Grunberg-Manago M, Putzer H. Condon C, et al. RNA. 2001 Feb;7(2):242-53. doi: 10.1017/s1355838201002163. RNA. 2001. PMID: 11233981 Free PMC article. - Chaperoning 5S RNA assembly.
Madru C, Lebaron S, Blaud M, Delbos L, Pipoli J, Pasmant E, Réty S, Leulliot N. Madru C, et al. Genes Dev. 2015 Jul 1;29(13):1432-46. doi: 10.1101/gad.260349.115. Genes Dev. 2015. PMID: 26159998 Free PMC article. - Use of mutant RNAs in studies on yeast 5S rRNA structure and function.
Nazar RN, Van Ryk DI, Lee Y, Guyer CD. Nazar RN, et al. Biochem Cell Biol. 1991 Apr;69(4):217-22. doi: 10.1139/o91-033. Biochem Cell Biol. 1991. PMID: 2054154 Review.
Cited by
- Optimization of ribosome structure and function by rRNA base modification.
Baxter-Roshek JL, Petrov AN, Dinman JD. Baxter-Roshek JL, et al. PLoS One. 2007 Jan 24;2(1):e174. doi: 10.1371/journal.pone.0000174. PLoS One. 2007. PMID: 17245450 Free PMC article. - The eukaryotic ribosome: current status and challenges.
Dinman JD. Dinman JD. J Biol Chem. 2009 May 1;284(18):11761-5. doi: 10.1074/jbc.R800074200. Epub 2008 Dec 31. J Biol Chem. 2009. PMID: 19117941 Free PMC article. Review. - Ribosome engineering reveals the importance of 5S rRNA autonomy for ribosome assembly.
Huang S, Aleksashin NA, Loveland AB, Klepacki D, Reier K, Kefi A, Szal T, Remme J, Jaeger L, Vázquez-Laslop N, Korostelev AA, Mankin AS. Huang S, et al. Nat Commun. 2020 Jun 9;11(1):2900. doi: 10.1038/s41467-020-16694-8. Nat Commun. 2020. PMID: 32518240 Free PMC article. - Specific effects of ribosome-tethered molecular chaperones on programmed -1 ribosomal frameshifting.
Muldoon-Jacobs KL, Dinman JD. Muldoon-Jacobs KL, et al. Eukaryot Cell. 2006 Apr;5(4):762-70. doi: 10.1128/EC.5.4.762-770.2006. Eukaryot Cell. 2006. PMID: 16607023 Free PMC article. - Heterochromatic siRNAs and DDM1 independently silence aberrant 5S rDNA transcripts in Arabidopsis.
Blevins T, Pontes O, Pikaard CS, Meins F Jr. Blevins T, et al. PLoS One. 2009 Jun 16;4(6):e5932. doi: 10.1371/journal.pone.0005932. PLoS One. 2009. PMID: 19529764 Free PMC article.
References
- Altschul SF, Gish W, Miller E, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4Å resolution. Science. 2000;289:905–920. - PubMed
- Betzel C, Lorenz S, Furste JP, Bald R, Zhang M, Schneider TR, Wilson KS, Erdmann VA. Crystal structure of domain A of Thermus flavus 5S rRNA and the contribution of water molecules to its structure. FEBS Lett. 1994;351:159–164. - PubMed
- Boeke JD, Xu H, Fink GR. A general method for the chromosomal amplification of genes into yeast. Science. 1988;239:280–282. - PubMed
- Bogdanov AA, Dontsova OA, Dokudovskaya SS, Lavrik IN. Structure and function of 5S rRNA in the ribosome. Biochem Cell Biol. 1995;73:869–876. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases