Functional polarity is introduced by Dicer processing of short substrate RNAs - PubMed (original) (raw)
Functional polarity is introduced by Dicer processing of short substrate RNAs
Scott D Rose et al. Nucleic Acids Res. 2005.
Abstract
Synthetic RNA duplexes that are substrates for Dicer are potent triggers of RNA interference (RNAi). Blunt 27mer duplexes can be up to 100-fold more potent than traditional 21mer duplexes. Not all 27mer duplexes show increased potency. Evaluation of the products of in vitro dicing reactions using electrospray ionization mass spectrometry reveals that a variety of products can be produced by Dicer cleavage. Use of asymmetric duplexes having a single 2-base 3'-overhang restricts the heterogeneity that results from dicing. Inclusion of DNA residues at the ends of blunt duplexes also limits heterogeneity. Combination of asymmetric 2-base 3'-overhang with 3'-DNA residues on the blunt end result in a duplex form which directs dicing to predictably yield a single primary cleavage product. It is therefore possible to design a 27mer duplex which is processed by Dicer to yield a specific, desired 21mer species. Using this strategy, two different 27mers can be designed that result in the same 21mer after dicing, one where the 3'-overhang resides on the antisense (AS) strand and dicing proceeds to the 'right' ('R') and one where the 3'-overhang resides on the sense (S) strand and dicing proceeds to the 'left' ('L'). Interestingly, the 'R' version of the asymmetric 27mer is generally more potent in reducing target gene levels than the 'L' version 27mer. Strand targeting experiments show asymmetric strand utilization between the two different 27mer forms, with the 'R' form favoring S strand and the 'L' form favoring AS strand silencing. Thus, Dicer processing confers functional polarity within the RNAi pathway.
Figures
Figure 1
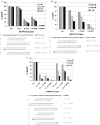
ESI-MS analysis of in vitro dicing reactions. Mass spectra of duplexes are shown before (top) and after (middle) digestion with recombinant human Dicer. Sequence of the substrate RNA duplex is provided with detected cleavage products along with calculated molecular weight and strand length (bottom). RNA bases are upper case, DNA bases are lower case bold and ‘p’ represents 5′-phosphate. (A) Blunt duplex EGFPS2 R 27/27; (B) asymmetric duplex EGFPS2 R 25/27; (C) asymmetric duplex EGFPS2 R 27/25; (D) asymmetric duplex EGFPS2 R 27/25(3′D); (E) asymmetric duplex EGFPS2 R 25D/27; (F) asymmetric duplex EGFPS2 L 27/25D; (G) asymmetric duplex EGFPS1 R 25D/27; (H) asymmetric duplex EGFPS1 L 27/25D.
Figure 1
ESI-MS analysis of in vitro dicing reactions. Mass spectra of duplexes are shown before (top) and after (middle) digestion with recombinant human Dicer. Sequence of the substrate RNA duplex is provided with detected cleavage products along with calculated molecular weight and strand length (bottom). RNA bases are upper case, DNA bases are lower case bold and ‘p’ represents 5′-phosphate. (A) Blunt duplex EGFPS2 R 27/27; (B) asymmetric duplex EGFPS2 R 25/27; (C) asymmetric duplex EGFPS2 R 27/25; (D) asymmetric duplex EGFPS2 R 27/25(3′D); (E) asymmetric duplex EGFPS2 R 25D/27; (F) asymmetric duplex EGFPS2 L 27/25D; (G) asymmetric duplex EGFPS1 R 25D/27; (H) asymmetric duplex EGFPS1 L 27/25D.
Figure 1
ESI-MS analysis of in vitro dicing reactions. Mass spectra of duplexes are shown before (top) and after (middle) digestion with recombinant human Dicer. Sequence of the substrate RNA duplex is provided with detected cleavage products along with calculated molecular weight and strand length (bottom). RNA bases are upper case, DNA bases are lower case bold and ‘p’ represents 5′-phosphate. (A) Blunt duplex EGFPS2 R 27/27; (B) asymmetric duplex EGFPS2 R 25/27; (C) asymmetric duplex EGFPS2 R 27/25; (D) asymmetric duplex EGFPS2 R 27/25(3′D); (E) asymmetric duplex EGFPS2 R 25D/27; (F) asymmetric duplex EGFPS2 L 27/25D; (G) asymmetric duplex EGFPS1 R 25D/27; (H) asymmetric duplex EGFPS1 L 27/25D.
Figure 1
ESI-MS analysis of in vitro dicing reactions. Mass spectra of duplexes are shown before (top) and after (middle) digestion with recombinant human Dicer. Sequence of the substrate RNA duplex is provided with detected cleavage products along with calculated molecular weight and strand length (bottom). RNA bases are upper case, DNA bases are lower case bold and ‘p’ represents 5′-phosphate. (A) Blunt duplex EGFPS2 R 27/27; (B) asymmetric duplex EGFPS2 R 25/27; (C) asymmetric duplex EGFPS2 R 27/25; (D) asymmetric duplex EGFPS2 R 27/25(3′D); (E) asymmetric duplex EGFPS2 R 25D/27; (F) asymmetric duplex EGFPS2 L 27/25D; (G) asymmetric duplex EGFPS1 R 25D/27; (H) asymmetric duplex EGFPS1 L 27/25D.
Figure 2
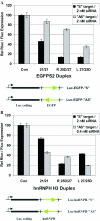
‘R’ form duplexes are more potent than ‘L’ form duplexes. Top: relative expression data are shown. Bottom: target sequences (S strand) are shown and transfected RNAi duplexes are aligned beneath in duplex form with sense strand top (5′→3′) and antisense strand bottom (3′→5′). RNA bases are upper case, DNA bases are lower case bold, and ‘p’ represents 5′-phosphate. (A) Relative EGFP fluorescence was measured following co-transfection of an EGFP expression plasmid and EGFPS2 RNA duplexes; (B) relative EGFP fluorescence was measured following co-transfection of an EGFP expression plasmid and EGFPS1 RNA duplexes; (C) relative EGFP fluorescence was measured following co-transfection of an EGFP-hnRNPH fusion vector and hnRNPH RNA duplexes; (D) relative light emission from firefly luciferase was measured following co-transfection of a luciferase expression vector and luciferase RNA duplexes; (E) RNA duplexes specific for La antigen mRNA were transfected into cells at 2.5 nM concentration and protein extracts were prepared 72 h post-transfection. Western blots were done using anti-La antibodies and anti-enolase (control) antibodies. RNA duplexes were transfected into cells at 1, 2.5 and 10 nM concentrations, and RNA was prepared 24 h post-transfection. Quantitative real-time RT–PCR was used to measure relative La mRNA levels, using RPLP0 as an internal normalization standard.
Figure 2
‘R’ form duplexes are more potent than ‘L’ form duplexes. Top: relative expression data are shown. Bottom: target sequences (S strand) are shown and transfected RNAi duplexes are aligned beneath in duplex form with sense strand top (5′→3′) and antisense strand bottom (3′→5′). RNA bases are upper case, DNA bases are lower case bold, and ‘p’ represents 5′-phosphate. (A) Relative EGFP fluorescence was measured following co-transfection of an EGFP expression plasmid and EGFPS2 RNA duplexes; (B) relative EGFP fluorescence was measured following co-transfection of an EGFP expression plasmid and EGFPS1 RNA duplexes; (C) relative EGFP fluorescence was measured following co-transfection of an EGFP-hnRNPH fusion vector and hnRNPH RNA duplexes; (D) relative light emission from firefly luciferase was measured following co-transfection of a luciferase expression vector and luciferase RNA duplexes; (E) RNA duplexes specific for La antigen mRNA were transfected into cells at 2.5 nM concentration and protein extracts were prepared 72 h post-transfection. Western blots were done using anti-La antibodies and anti-enolase (control) antibodies. RNA duplexes were transfected into cells at 1, 2.5 and 10 nM concentrations, and RNA was prepared 24 h post-transfection. Quantitative real-time RT–PCR was used to measure relative La mRNA levels, using RPLP0 as an internal normalization standard.
Figure 3
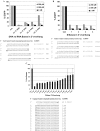
Strand bias is introduced by the direction of Dicer processing. Reporter constructs were made in the psiCheck-2™ vector that contained a fragment of EGFP or hnRNPH coding sequence cloned into the 3′-UTR of the Renilla luciferase gene in both ‘S’ and ‘AS’ orientations. Reporter plasmids were co-transfected into cells without (control) or with the indicated RNA duplexes and luciferase activity was measured 24 h post-transfection. Results are reported as the ratio of Renilla luciferase (target) to firefly luciferase (internal control); (A) EGFP strand targeting; (B) hnRNPH strand targeting. Schematic diagrams of the four synthetic reporter constructs employed with orientation of target sequences relative to Renilla luciferase are shown.
Figure 4
Structure–activity features of Dicer–substrate RNA duplexes. (A) Comparison of activity of ‘R’ form 27mer duplexes with RNA versus DNA bases in the single 2-base 3′-overhang. (B) Comparison of activity of ‘R’ form 27mer duplexes with 1, 2, 3 or 4-base RNA 3′-overhangs. (C) Comparison of activity of ‘R’ form 27mer duplexes with every possible RNA 2-base 3′-overhang. Top: For A and B, an EGFP-hnRNPH fusion protein expression plasmid was transfected into cells without (control) or with the indicated RNA duplexes and EGFP fluorescence was measured at 48 h post-transfection. For C, a luciferase expression vector containing hnRNPH sequence in the 3′-UTR was transfected into cells without (control) or with the indicated RNA duplexes and luciferase activity was assayed 48 h post-transfection. Bottom: hnRNPH target sequence (S strand) is shown and duplexes employed in transfection are aligned beneath in duplex form with sense strand top (5′→3′) and antisense strand bottom (3′→5′). RNA bases are upper case, DNA bases are lower case bold, and ‘p’ represents 5′-phosphate. Two neighboring sites were targeted, H-1 and H-3 shifted by two bases along the hnRNPH sequence.
Similar articles
- Deep Sequencing Analyses of DsiRNAs Reveal the Influence of 3' Terminal Overhangs on Dicing Polarity, Strand Selectivity, and RNA Editing of siRNAs.
Zhou J, Song MS, Jacobi AM, Behlke MA, Wu X, Rossi JJ. Zhou J, et al. Mol Ther Nucleic Acids. 2012 Apr 3;1(4):e17. doi: 10.1038/mtna.2012.6. Mol Ther Nucleic Acids. 2012. PMID: 23343928 Free PMC article. - Short interfering RNA strand selection is independent of dsRNA processing polarity during RNAi in Drosophila.
Preall JB, He Z, Gorra JM, Sontheimer EJ. Preall JB, et al. Curr Biol. 2006 Mar 7;16(5):530-5. doi: 10.1016/j.cub.2006.01.061. Curr Biol. 2006. PMID: 16527750 - Continuous fluorescence-based method for assessing dicer cleavage efficiency reveals 3' overhang nucleotide preference.
DiNitto JP, Wang L, Wu JC. DiNitto JP, et al. Biotechniques. 2010 Apr;48(4):303-11. doi: 10.2144/000113402. Biotechniques. 2010. PMID: 20569207 - Approaches for chemically synthesized siRNA and vector-mediated RNAi.
Amarzguioui M, Rossi JJ, Kim D. Amarzguioui M, et al. FEBS Lett. 2005 Oct 31;579(26):5974-81. doi: 10.1016/j.febslet.2005.08.070. Epub 2005 Sep 20. FEBS Lett. 2005. PMID: 16199038 Review. - Dicer-independent processing of small RNA duplexes: mechanistic insights and applications.
Herrera-Carrillo E, Berkhout B. Herrera-Carrillo E, et al. Nucleic Acids Res. 2017 Oct 13;45(18):10369-10379. doi: 10.1093/nar/gkx779. Nucleic Acids Res. 2017. PMID: 28977573 Free PMC article. Review.
Cited by
- The Application of Light-Assisted Drying to the Thermal Stabilization of Nucleic Acid Nanoparticles.
Anh Lam P, Furr DP, Tran A, McKeough RQ, Beasock D, Chandler M, Afonin KA, Trammell SR. Anh Lam P, et al. Biopreserv Biobank. 2022 Oct;20(5):451-460. doi: 10.1089/bio.2022.0035. Epub 2022 Sep 2. Biopreserv Biobank. 2022. PMID: 36067075 Free PMC article. - Progress towards in vivo use of siRNAs.
Behlke MA. Behlke MA. Mol Ther. 2006 Apr;13(4):644-70. doi: 10.1016/j.ymthe.2006.01.001. Epub 2006 Feb 14. Mol Ther. 2006. PMID: 16481219 Free PMC article. Review. - Sequence-non-specific effects of RNA interference triggers and microRNA regulators.
Olejniczak M, Galka P, Krzyzosiak WJ. Olejniczak M, et al. Nucleic Acids Res. 2010 Jan;38(1):1-16. doi: 10.1093/nar/gkp829. Epub 2009 Oct 20. Nucleic Acids Res. 2010. PMID: 19843612 Free PMC article. Review. - Nanoscale metal-organic frameworks for the delivery of nucleic acids to cancer cells.
Li X, Chandler M, Avila YI, Arroyo-Becker SI, Patriarche G, Vargas-Berenguel A, Casas-Solvas JM, Afonin KA, Gref R. Li X, et al. Int J Pharm X. 2023 Jan 30;5:100161. doi: 10.1016/j.ijpx.2023.100161. eCollection 2023 Dec. Int J Pharm X. 2023. PMID: 36817971 Free PMC article. - Differences in silencing of mismatched targets by sliced versus diced siRNAs.
Sun G, Wang J, Huang Y, Yuan CW, Zhang K, Hu S, Chen L, Lin RJ, Yen Y, Riggs AD. Sun G, et al. Nucleic Acids Res. 2018 Jul 27;46(13):6806-6822. doi: 10.1093/nar/gky287. Nucleic Acids Res. 2018. PMID: 29718312 Free PMC article.
References
- Kim D.H., Behlke M.A., Rose S.D., Chang M.S., Choi S., Rossi J.J. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2005;23:222–226. - PubMed
- Mello C.C., Conte D., Jr Revealing the world of RNA interference. Nature. 2004;431:338–342. - PubMed
- Meister G., Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. - PubMed
- Hannon G.J., Rossi J.J. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI29329/AI/NIAID NIH HHS/United States
- R01 HL074704/HL/NHLBI NIH HHS/United States
- AI42552/AI/NIAID NIH HHS/United States
- R37 AI029329/AI/NIAID NIH HHS/United States
- HL074704/HL/NHLBI NIH HHS/United States
- R01 AI042552/AI/NIAID NIH HHS/United States
- R01 AI029329/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials