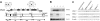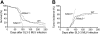A tumor-suppressor function for NFATc3 in T-cell lymphomagenesis by murine leukemia virus - PubMed (original) (raw)
. 2005 Nov 15;106(10):3546-52.
doi: 10.1182/blood-2005-02-0493. Epub 2005 Jul 28.
Annette Balle Sørensen, Mindaugas Andrulis, Bruce Wang, Eisaku Kondo, Randi Jessen, Laszlo Krenacs, Eva Stelkovics, Matthias Wabl, Edgar Serfling, Alois Palmetshofer, Finn Skou Pedersen
Affiliations
- PMID: 16051745
- PMCID: PMC1895049
- DOI: 10.1182/blood-2005-02-0493
A tumor-suppressor function for NFATc3 in T-cell lymphomagenesis by murine leukemia virus
Sys Zoffmann Glud et al. Blood. 2005.
Abstract
Nuclear factor of activated T cell (NFAT) transcription factors play a central role in differentiation, activation, and elimination of lymphocytes. We here report on the finding of provirus integration into the Nfatc3 locus in T-cell lymphomas induced by the murine lymphomagenic retrovirus SL3-3 and show that NFATc3 expression is repressed in these lymphomas. The provirus insertions are positioned close to the Nfatc3 promoter or a putative polyadenylated RNA (polyA) region. Furthermore, we demonstrate that NFATc3-deficient mice infected with SL3-3 develop T-cell lymphomas faster and with higher frequencies than wild-type mice or NFATc2-deficient mice. These results identify NFATc3 as a tumor suppressor for the development of murine T-cell lymphomas induced by the retrovirus SL3-3.
Figures
Figure 1.
Insertion of retrovirus SL3-3 in the Nfatc3 locus represses NFATc3 expression. (A) Schematic representation of Nfatc3 with SL3-3 MLV insertion sites and orientations indicated by arrowheads (▸). The numbers adjacent to the proviruses denote the distance (in bp) between provirus insertions and the upstream and downstream exons or the distance between neighboring insertions. TAD indicates transactivation domain; NLS, nuclear localization signal; a, calcineurin binding site; b, region within the regulatory domain (RD) containing multiple phosphorylation sites; c, the DNA-binding motif in the DNA-binding domain (DBD); TAD/NES, combinatorial C-terminal TAD (TAD-C) and nuclear export signal found in NFATc1 and NFATc3; □, the alternative 33-aa sequence of the NFATx2 isoform of NFATc3 lacking TAD-C motif; ▵, the 30-aa deletion of an inactive isoform of NFATc3; and *, location of a putative polyA site (AATAAATAGTCTTTTTT) in NFATc3 with start at position chr8:105418773. (B) Western blot analysis of protein extracts from lymphomas with SL3-3 insertion within the Nfatc3 locus using polyclonal anti-NFATc1 or anti-NFATc3 antibody for staining (“Materials and methods”). Protein extracts from normal thymus of a wild-type mouse (lane 1) are compared with lysates from tumors of _Nfatc3_-/- (lane 2) or wild-type mice without (lane 3) and with (lanes 4 and 5, representing tumors with insertion in intron 8 and at 1.6-kb upstream, respectively) SL3-3 MLV insertion at the Nfatc3 locus. (C) RNA expression of NFAT genes in SL3-3–induced tumors as determined by semiquantitative RT-PCR. Shown are the expression in normal thymus, thymic tumors derived from wild-type, _Nfatc2_-/--, and _Nfatc3_-/- mice, and thymic tumors with provirus insertion in Nfatc3 locus (at intron 8 and 1.6-kb upstream). cDNA amounts were equalized to β-actin signals. WT indicates wild type.
Figure 2.
Diminished survival and increased tumor incidence in _Nfatc3_-/- mice after SL3-3 infection. (A) Survival curves of _Nfatc3_-/- (n = 16/16), _Nfatc2_-/- (n = 80/88), or wild-type (n = 48/58) (including heterozygotes) BALB/c mice infected with SL3-3. (B) The cumulative incidence of tumor development of _Nfatc3_-/- (n = 14/16), _Nfatc2_-/- (n = 53/88), and wild-type (n = 38/58) mice based on enlarged lymphoid organs as defined previously (for details, see “Materials and methods”).
Figure 3.
Immunohistochemical analyses of SL3-3–induced tumors. (A) Sections of a representative tumor of the thymus stained with either H&E or antibodies against CD3ε (CD3), TdT, and myeloperoxidase (MPO). (B) Immunohistochemical stainings of CD3ε+ tumors from different animals for double fluorescent analysis of CD4 and CD8 expression. All of the samples shown were either CD3ε bright (nos. 160, 163, 407 all thymus) or CD3ε dim (no. 275; mesentery lymph node). The genotypes of the mice with the tumors shown are _Nfatc3_-/- (nos. 160, 163, 407) and _Nfatc2_-/- (no. 275). Images were acquired with Leica TCS SP2 Confocal System equipped with a Leica DMRE microscope with an HCX PL APO 40×/1.25 NA oil CS objective lens. Images were processed with Leica Confocal Software version 2.61 (all from Leica Microsystems, Mannheim, Germany).
Figure 4.
SL3-3 infection leads to various subtypes of T-cell lymphomas in wild-type and NFATc2- or NFATc3-deficient mice. Flow cytometry analysis of cells from (A) spleen of mock-infected _Nfatc3_-/- mice, (B) spleen lymphoma from a SL3-3–infected _Nfatc3_-/- mouse, (C) thymus lymphoma from an SL3-3–infected wild-type mouse, and (D) spleen lymphoma from an SL3-3–infected _Nfatc2_-/- mouse. (A-B) To distinguish between T cells, myeloid cells, and B cells, cells were stained with a mixture of anti-CD3–PE, anti-CD11b–FITC, and anti-CD45/B220–PerCP Abs, respectively. In panels C and D, a mixture of anti-CD3–PE, anti-CD4–FITC, and anti-CD8–PerCP–Cy5.5 antibodies was used to distinguish among T-cell subtypes. Two different populations of T cells in panel D are indicated with rectangles 1 and 2 representing CD3+CD4+CD8+ cells and CD3+CD4-(dim)CD8+(dim) cells, respectively. Note that parallel analysis of both thymus and spleen tumors derived from the same animal revealed basically identical FACS profiles in most of the cases investigated. Five percent probability contour plots are shown. Grids are positioned according to background fluorescence of isotype controls. Numbers in quadrants indicate the percentage of cells within the limits of the quadrants. For a summary of tumor phenotypes, see Table 2.
Similar articles
- Stability of AML1 (core) site enhancer mutations in T lymphomas induced by attenuated SL3-3 murine leukemia virus mutants.
Amtoft HW, Sørensen AB, Bareil C, Schmidt J, Luz A, Pedersen FS. Amtoft HW, et al. J Virol. 1997 Jul;71(7):5080-7. doi: 10.1128/JVI.71.7.5080-5087.1997. J Virol. 1997. PMID: 9188573 Free PMC article. - Sint1, a common integration site in SL3-3-induced T-cell lymphomas, harbors a putative proto-oncogene with homology to the septin gene family.
Sørensen AB, Lund AH, Ethelberg S, Copeland NG, Jenkins NA, Pedersen FS. Sørensen AB, et al. J Virol. 2000 Mar;74(5):2161-8. doi: 10.1128/jvi.74.5.2161-2168.2000. J Virol. 2000. PMID: 10666245 Free PMC article. - The role of the immune response in MULV-induced lymphomagenesis.
Marshall DJ, Gaulton GN. Marshall DJ, et al. Leukemia. 1996 Dec;10(12):1860-6. Leukemia. 1996. PMID: 8946922 Review. - AID for innate immunity to retroviral transformation.
Schlissel MS, Kuo TC. Schlissel MS, et al. Immunity. 2006 Jun;24(6):671-672. doi: 10.1016/j.immuni.2006.06.005. Immunity. 2006. PMID: 16782023 Review.
Cited by
- Nuclear factor of activated T-cell c3 inhibition of mammalian target of rapamycin signaling through induction of regulated in development and DNA damage response 1 in human intestinal cells.
Zhou Y, Wang Q, Guo Z, Weiss HL, Evers BM. Zhou Y, et al. Mol Biol Cell. 2012 Aug;23(15):2963-72. doi: 10.1091/mbc.E12-01-0037. Epub 2012 Jun 13. Mol Biol Cell. 2012. PMID: 22696685 Free PMC article. - AucPR: an AUC-based approach using penalized regression for disease prediction with high-dimensional omics data.
Yu W, Park T. Yu W, et al. BMC Genomics. 2014;15 Suppl 10(Suppl 10):S1. doi: 10.1186/1471-2164-15-S10-S1. Epub 2014 Dec 12. BMC Genomics. 2014. PMID: 25559769 Free PMC article. - Herpesviral G protein-coupled receptors activate NFAT to induce tumor formation via inhibiting the SERCA calcium ATPase.
Zhang J, He S, Wang Y, Brulois K, Lan K, Jung JU, Feng P. Zhang J, et al. PLoS Pathog. 2015 Mar 26;11(3):e1004768. doi: 10.1371/journal.ppat.1004768. eCollection 2015 Mar. PLoS Pathog. 2015. PMID: 25811856 Free PMC article. - Epigenetic changes and suppression of the nuclear factor of activated T cell 1 (NFATC1) promoter in human lymphomas with defects in immunoreceptor signaling.
Akimzhanov A, Krenacs L, Schlegel T, Klein-Hessling S, Bagdi E, Stelkovics E, Kondo E, Chuvpilo S, Wilke P, Avots A, Gattenlöhner S, Müller-Hermelink HK, Palmetshofer A, Serfling E. Akimzhanov A, et al. Am J Pathol. 2008 Jan;172(1):215-24. doi: 10.2353/ajpath.2008.070294. Epub 2007 Dec 21. Am J Pathol. 2008. PMID: 18156209 Free PMC article. - An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors.
Dong B, Kim S, Hong S, Das Gupta J, Malathi K, Klein EA, Ganem D, Derisi JL, Chow SA, Silverman RH. Dong B, et al. Proc Natl Acad Sci U S A. 2007 Jan 30;104(5):1655-60. doi: 10.1073/pnas.0610291104. Epub 2007 Jan 18. Proc Natl Acad Sci U S A. 2007. PMID: 17234809 Free PMC article.
References
- Serfling E, Berberich-Siebelt F, Chuvpilo S, et al. The role of NF-AT transcription factors in T cell activation and differentiation. Biochim Biophys Acta. 2000;1498: 1-18. - PubMed
- Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109(suppl 2): S67-S79. - PubMed
- Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17: 2205-2232. - PubMed
- Peng SL, Gerth AJ, Ranger AM, Glimcher LH. NFATc1 and NFATc2 together control both T and B cell activation and differentiation. Immunity. 2001;14: 13-20. - PubMed
- Xanthoudakis S, Viola JP, Shaw KT, et al. An enhanced immune response in mice lacking the transcription factor NFAT1. Science. 1996;272: 892-895. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous