HLA-B63 presents HLA-B57/B58-restricted cytotoxic T-lymphocyte epitopes and is associated with low human immunodeficiency virus load - PubMed (original) (raw)
. 2005 Aug;79(16):10218-25.
doi: 10.1128/JVI.79.16.10218-10225.2005.
Sharon Adams, Photini Kiepiela, Caitlyn H Linde, Hannah S Hewitt, Mathias Lichterfeld, Kaori Sango, Nancy V Brown, Eunice Pae, Alysse G Wurcel, Marcus Altfeld, Margaret E Feeney, Todd M Allen, Timothy Roach, M Anne St John, Eric S Daar, Eric Rosenberg, Bette Korber, Francesco Marincola, Bruce D Walker, Philip J R Goulder, Christian Brander
Affiliations
- PMID: 16051815
- PMCID: PMC1182636
- DOI: 10.1128/JVI.79.16.10218-10225.2005
HLA-B63 presents HLA-B57/B58-restricted cytotoxic T-lymphocyte epitopes and is associated with low human immunodeficiency virus load
Nicole Frahm et al. J Virol. 2005 Aug.
Abstract
Several HLA class I alleles have been associated with slow human immunodeficiency virus (HIV) disease progression, supporting the important role HLA class I-restricted cytotoxic T lymphocytes (CTL) play in controlling HIV infection. HLA-B63, the serological marker for the closely related HLA-B*1516 and HLA-B*1517 alleles, shares the epitope binding motif of HLA-B57 and HLA-B58, two alleles that have been associated with slow HIV disease progression. We investigated whether HIV-infected individuals who express HLA-B63 generate CTL responses that are comparable in breadth and specificity to those of HLA-B57/58-positive subjects and whether HLA-B63-positive individuals would also present with lower viral set points than the general population. The data show that HLA-B63-positive individuals indeed mounted responses to previously identified HLA-B57-restricted epitopes as well as towards novel, HLA-B63-restricted CTL targets that, in turn, can be presented by HLA-B57 and HLA-B58. HLA-B63-positive subjects generated these responses early in acute HIV infection and were able to control HIV replication in the absence of antiretroviral treatment with a median viral load of 3,280 RNA copies/ml. The data support an important role of the presented epitope in mediating relative control of HIV replication and help to better define immune correlates of controlled HIV infection.
Figures
FIG. 1.
Epitope fine mapping of HLA-B57-restricted CTL epitopes in HLA-B63-positive subjects. Responses to serially 10-fold-diluted, truncated peptides in ELISPOT are shown for epitopes KF11 (a), KY10 (b), and IW9 (c, d). The optimal epitope was defined as the shortest epitope inducing the strongest response at the lowest concentration (4). Captions include the subjects and their respective HLA types.
FIG. 2.
Epitope fine mapping of HLA-B63-restricted CTL epitopes. Responses to serially 10-fold-diluted, truncated peptides in ELISPOT are shown for epitopes LL9 (OLP 85) (a, b), RY11 (OLP 11) (c), YY9 (OLP 84) (d), FF9 (OLP 23) (e), and KV8 (OLP 162) (f). The optimal epitope was defined as the shortest epitope inducing the strongest response at the lowest concentration (4). Captions include the subjects and their respective HLA types, and used truncations are shown in the legend.
FIG. 3.
Magnitude of responses during acute infection. The relative magnitude of responses restricted by HLA-B63 or other alleles is shown for subjects B19 (a) and B20 (b), indicating the preferential targeting of HLA-B63 responses early in infection.
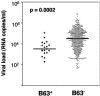
FIG. 4.
Viral loads of HLA-B63-positive subjects are compared to the remainder of the cohorts (including B57/58-positive subjects). Viral set points are shown for 19 untreated, chronically HIV clade B- or C-infected, HLA-B63-positive individuals (large diamonds) and 576 untreated, HLA-B63-negative clade B- and clade C-infected individuals (small diamonds). Medians are shown as horizontal black lines.
Similar articles
- Characterization of HLA-B57-restricted human immunodeficiency virus type 1 Gag- and RT-specific cytotoxic T lymphocyte responses.
Klein MR, van der Burg SH, Hovenkamp E, Holwerda AM, Drijfhout JW, Melief CJ, Miedema F. Klein MR, et al. J Gen Virol. 1998 Sep;79 ( Pt 9):2191-201. doi: 10.1099/0022-1317-79-9-2191. J Gen Virol. 1998. PMID: 9747728 - Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection.
Altfeld M, Addo MM, Rosenberg ES, Hecht FM, Lee PK, Vogel M, Yu XG, Draenert R, Johnston MN, Strick D, Allen TM, Feeney ME, Kahn JO, Sekaly RP, Levy JA, Rockstroh JK, Goulder PJ, Walker BD. Altfeld M, et al. AIDS. 2003 Dec 5;17(18):2581-91. doi: 10.1097/00002030-200312050-00005. AIDS. 2003. PMID: 14685052 - Accumulation of Pol Mutations Selected by HLA-B*52:01-C*12:02 Protective Haplotype-Restricted Cytotoxic T Lymphocytes Causes Low Plasma Viral Load Due to Low Viral Fitness of Mutant Viruses.
Murakoshi H, Koyanagi M, Chikata T, Rahman MA, Kuse N, Sakai K, Gatanaga H, Oka S, Takiguchi M. Murakoshi H, et al. J Virol. 2017 Jan 31;91(4):e02082-16. doi: 10.1128/JVI.02082-16. Print 2017 Feb 15. J Virol. 2017. PMID: 27903797 Free PMC article. - Identification of highly conserved and broadly cross-reactive HIV type 1 cytotoxic T lymphocyte epitopes as candidate immunogens for inclusion in Mycobacterium bovis BCG-vectored HIV vaccines.
Ferrari G, Kostyu DD, Cox J, Dawson DV, Flores J, Weinhold KJ, Osmanov S. Ferrari G, et al. AIDS Res Hum Retroviruses. 2000 Sep 20;16(14):1433-43. doi: 10.1089/08892220050140982. AIDS Res Hum Retroviruses. 2000. PMID: 11018863 Review. - Protective HLA-B57: T cell and natural killer cell recognition in HIV infection.
Lobos CA, Downing J, D'Orsogna LJ, Chatzileontiadou DSM, Gras S. Lobos CA, et al. Biochem Soc Trans. 2022 Oct 31;50(5):1329-1339. doi: 10.1042/BST20220244. Biochem Soc Trans. 2022. PMID: 36111814 Free PMC article. Review.
Cited by
- A statistical framework for modeling HLA-dependent T cell response data.
Listgarten J, Frahm N, Kadie C, Brander C, Heckerman D. Listgarten J, et al. PLoS Comput Biol. 2007 Oct;3(10):1879-86. doi: 10.1371/journal.pcbi.0030188. PLoS Comput Biol. 2007. PMID: 17937494 Free PMC article. - HLA class I-driven evolution of human immunodeficiency virus type 1 subtype c proteome: immune escape and viral load.
Rousseau CM, Daniels MG, Carlson JM, Kadie C, Crawford H, Prendergast A, Matthews P, Payne R, Rolland M, Raugi DN, Maust BS, Learn GH, Nickle DC, Coovadia H, Ndung'u T, Frahm N, Brander C, Walker BD, Goulder PJ, Bhattacharya T, Heckerman DE, Korber BT, Mullins JI. Rousseau CM, et al. J Virol. 2008 Jul;82(13):6434-46. doi: 10.1128/JVI.02455-07. Epub 2008 Apr 23. J Virol. 2008. PMID: 18434400 Free PMC article. - Use of RT-defective HIV virions: new tool to evaluate specific response in chronic asymptomatic HIV-infected individuals.
Guardo AC, Alvarez-Fernández C, Arberas H, García-Pérez J, García F, Bargalló ME, Maleno MJ, Gatell JM, Mothe B, Alcami J, Sánchez-Palomino S, Plana M. Guardo AC, et al. PLoS One. 2013;8(3):e58927. doi: 10.1371/journal.pone.0058927. Epub 2013 Mar 14. PLoS One. 2013. PMID: 23516578 Free PMC article. - Aminopeptidase substrate preference affects HIV epitope presentation and predicts immune escape patterns in HIV-infected individuals.
Zhang SC, Martin E, Shimada M, Godfrey SB, Fricke J, Locastro S, Lai NY, Liebesny P, Carlson JM, Brumme CJ, Ogbechie OA, Chen H, Walker BD, Brumme ZL, Kavanagh DG, Le Gall S. Zhang SC, et al. J Immunol. 2012 Jun 15;188(12):5924-34. doi: 10.4049/jimmunol.1200219. Epub 2012 May 14. J Immunol. 2012. PMID: 22586036 Free PMC article. - HLA alleles associated with slow progression to AIDS truly prefer to present HIV-1 p24.
Borghans JA, Mølgaard A, de Boer RJ, Keşmir C. Borghans JA, et al. PLoS One. 2007 Sep 19;2(9):e920. doi: 10.1371/journal.pone.0000920. PLoS One. 2007. PMID: 17878955 Free PMC article.
References
- Allen, T. M., M. Altfeld, X. G. Yu, K. M. O'Sullivan, M. Lichterfeld, S. Le Gall, M. John, B. R. Mothe, P. K. Lee, E. T. Kalife, D. E. Cohen, K. A. Freedberg, D. A. Strick, M. N. Johnston, A. Sette, E. S. Rosenberg, S. A. Mallal, P. J. Goulder, C. Brander, and B. D. Walker. 2004. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J. Virol. 78:7069-7078. - PMC - PubMed
- Altfeld, M., M. M. Addo, E. S. Rosenberg, F. M. Hecht, P. K. Lee, M. Vogel, X. G. Yu, R. Draenert, M. N. Johnston, D. Strick, T. M. Allen, M. E. Feeney, J. O. Kahn, R. P. Sekaly, J. A. Levy, J. K. Rockstroh, P. J. Goulder, and B. D. Walker. 2003. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 17:2581-2591. - PubMed
- Barber, L. D., L. Percival, K. L. Arnett, J. E. Gumperz, L. Chen, and P. Parham. 1997. Polymorphism in the alpha 1 helix of the HLA-B heavy chain can have an overriding influence on peptide-binding specificity. J. Immunol. 158:1660-1669. - PubMed
- Brander, C., and B. D. Walker. 1996. The HLA class I restricted CTL response in HIV infection: systematic identification of optimal epitopes, p. IV50-IV60. In C. B. B. Korber, B. Walker, R. Koup, J. Moore, B. Haynes, and G. Meyers (ed.), HIV molecular immunology database. Theoretical biology and biophysics. Los Alamos National Laboratory, Los Alamos, N.M.
- Cao, K., J. Hollenbach, X. Shi, W. Shi, M. Chopek, and M. A. Fernandez-Vina. 2001. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum. Immunol. 62:1009-1030. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI-043638/AI/NIAID NIH HHS/United States
- N01-AI-15422/AI/NIAID NIH HHS/United States
- U01 AI043638/AI/NIAID NIH HHS/United States
- AI-067077/AI/NIAID NIH HHS/United States
- R01 AI067077/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials