A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons - PubMed (original) (raw)
FIG. 1.
Construction and analysis of _UBR1_−/− _UBR2_−/− cells. (A) The mammalian N-end rule pathway. N-terminal residues are indicated by single-letter abbreviations for amino acids. The ovals denote the rest of a protein substrate. (B) Southern analysis of UBR1 and UBR2 using wild-type (wt) and _UBR1_−/− _UBR2_−/− embryos with cDNA probes (46, 47). To determine UBR1 genotypes, DNAs from EFs were digested with SphI (5′ probe) or BamHI (3′ probe). To determine UBR2 genotypes, embryonic DNAs were digested with NheI (5′ probe) or BamHI/SalI (3′ probe). (C) Northern analysis of UBR1 and UBR2 using total RNAs from UBR1 +/− _UBR2_−/− and _UBR1_−/− _UBR2_−/− EFs. Different UBR1 and UBR2 cDNA fragments were used to probe the deleted regions in the knockout alleles (deleted) or both the deleted regions and its flanking regions (flanking) as described (46, 47). The two different UBR2 probes detected no significant UBR2 mRNA in _UBR1_−/− _UBR2_−/− EFs, while the UBR1 probe, which encompasses both the deleted region and its 3′-flanking region, detected _UBR1_-specific transcripts in _UBR1_−/− UBR2_−/− EFs. These aberrant transcripts appear to be the abnormally spliced RNAs transcribed from the P_UBR1 promoter. (D) Design of bead-conjugated synthetic peptides bearing different N-terminal amino acids. N-terminal residues of bead-conjugated peptide are indicated by three-letter abbreviations. An 11-mer peptide was covalently linked to a bead as described in Materials and Methods. (E) EF cytoplasmic proteins captured by a bead-conjugated Phe-peptide were separated in SDS-PAGE (5% Tris-glycine gel) and analyzed by immunoblotting (IB) using anti-UBR1 and UBR2 antibodies. The bottom panel shows immunoblotting analysis of actin for an input control. (F) Primary wild-type and _UBR1_−/− _UBR2_−/− EF cells were immortalized to increase transfection efficiency, and the resulting immortalized cells were transiently transfected with pcDNA3flagDHFRhaUb-X-nsP4flag (where X is Met, Arg, or Tyr) expressing fDHFRh-UbR48-X-nsP4f, a UPR-based fusion yielding cotranslationally the fDHFRh-UbR48 reference protein (denoted fDUb on the left) and X-nsP4f test protein (denoted X-nsP4 on the left). Cells were labeled for 12 min with [35S]methionine/cysteine, followed by a chase for 0, 1, and 2 h in the presence of cycloheximide, preparation of extracts, immunoprecipitation with anti-Flag M2-agarose (Sigma), SDS-PAGE, autoradiography, and quantitation, essentially as described (46). (G) Quantitation of the patterns in F using PhosphorImager. The amounts of 35S in an X-nsP4f, relative to 35S in the fDHFRh-UbR48 reference protein at the same time points, were plotted as percentages of this ratio for Met-nsP4f (bearing a stabilizing N-terminal residue) at time zero. Open and solid symbols denote data obtained with wild-type and _UBR1_−/− _UBR2_−/− extracts, respectively. ○, Met-nsP4; ▵, Tyr-nsP4; □, Arg-nsP4.
FIG. 2.
Identification of mouse N-recognins. (A) Peptide-pulldown assay using testis extracts and bead-conjugated peptides. Captured proteins were separated and visualized using silver staining. The identities of four major bands that are specifically and reproducibly captured by Phe- and/or Arg-peptide beads were determined using peptide mass fingerprinting. (B) UBR1, UBR2, and UBR4 bound from testis extract were eluted from Phe-peptide beads by the Phe-Ala dipeptide but not by Gly-Ala. (C) The amount of UBR4 bound by Phe-peptide beads from EF cell extracts does not depend strongly on the presence of UBR1 and/or UBR2. Proteins bound by either mock or Phe-peptide beads are indicated on the left with short bars: white, UBR1; gray, UBR2; and black, UBR4. (D) Testis proteins bound to bead-conjugated peptides bearing different N-terminal amino acids were immunoblotted with antibodies against UBR1, UBR2, UBR4, and UBR5. Mock, beads without peptide conjugation. (E) Semipurification of endogenous UBR1/UBR2 from an EF cytoplasmic extract using a peptide-pulldown assay. The precipitates/Phe-peptide beads complex prepared from cytoplasmic EF extracts were separated on SDS-PAGE and transferred onto a polyvinylidene difluoride membrane, followed by staining with Coomassie brilliant blue R-250 (Bio-Rad). Anti-UBR1 and -UBR2 immunoblotting confirmed enrichment of UBR1 and UBR2 (data not shown). (F) Northern blot analysis of UBR4 and β-actin using different mouse adult tissues. Total RNA (20 μg) was loaded in each lane. Ethidium bromide-stained ribosomal RNAs are shown as a loading control (bottom panel).
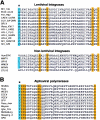
FIG. 3.
(A) Sequence alignment of the UBR box motifs. Conserved Cys and His residues are highlighted (cyan). Indicated by orange highlight is the Cys residue of Arabidopsis BIG, whose missense mutation perturbs auxin transport (24). Indicated by pink are the residues of S. cerevisiae UBR1 essential for degradation of type 1 N-end rule substrates (A. Webster, M. Ghislain, and A. Varshavsky, unpublished data). Note that different organisms contain different sets of UBR box proteins. Prefixes: m, Mus musculus; d, Drosophila melanogaster; a, Arabidopsis thaliana; sc, Saccharomyces cerevisiae; sp, Schizosaccharomyces pombe, c, Caenorhabditis elegans. Protein sequences listed are m-UBR1 (NP_033487), m-UBR2 (AAQ17202), m-UBR3 (T. Tasaki, Y. T. Kwon, and A. Varshavsky, unpublished data), m-UBR4 (this study), m-UBR5 (AAT28194), m-UBR6 (AAH55343), m-UBR7 (BAC40212), d-UBR1 (NP_573184), d-UBR3 (NP_572426), d-UBR4 (NP_476986), d-UBR5 (P51592), d-UBR6 (AAF54459), d-UBR7 (AAF53607), c-UBR1 (AAB42328), c-UBR3 (AAO38656), c-UBR4 (AAD47122), c-UBR5 (NP_492389), c-UBR6 (AAC48052), c-UBR7 (CAA99920), a-UBR1 (NP_195851), a-UBR4 (NP_186875), a-UBR7 (T08905), sc-UBR1 (P19812), sc-UBR2 (NP_013124), sp-UBR1 (CAB10108), sp-UBR3 (O60152), and sp-UBR7 (Q09329). (B) Locations of the UBR boxes and domains characteristic, in particular, of E3 Ub ligases in the mouse UBR1 to UBR7 proteins. UBR, UBR box; RING, RING finger; CRD, cysteine-rich domain; HECT, HECT domain; PHD, plant homeodomain. (C) Phylogenetic relationships of UBR box proteins. The relative evolutional distances between each UBR box sequence were calculated by a neighbor-joining method. The tree is unrooted and the length of each branch is proportional.
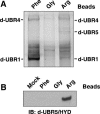
FIG. 4.
N-recognins in Drosophila melanogaster. (A) Peptide-pulldown analysis of Drosophila proteins in D.Mel-2 cells. Cytoplasmic proteins were precipitated by a bead-conjugated peptide bearing N-terminal Arg, Phe, or Gly. (B) Peptide-pulldown analysis followed by Western blot using anti-Drosophila UBR5/HYD antiserum.
FIG. 5.
Analysis of the in vivo degradation of Sindbis virus RNA polymerases bearing N-terminal Tyr in wild-type, _UBR4_RNAi, _UBR1_−/− _UBR2_−/− _UBR4_RNAi, and _UBR1_−/− _UBR2_−/− _Luc_RNAi stable cell lines. (A) Immunoblot analysis of UBR4 in _UBR4_RNAi, _UBR1_−/− _UBR2_−/− _UBR4_RNAi, and _UBR1_−/− _UBR2_−/− _Luc_RNAi stable cell lines. Wild-type (wt) and _UBR_−/− _UBR2_−/− EFs stably expressing UBR4 short-hairpin RNA (shRNA) were analyzed by immunoblot using anti-UBR4 antibody. Cells were transduced by recombinant retroviruses with four different short-hairpin RNA sequences (A, B, C, and D) for UBR4 RNAi or firefly luciferase (Luc) and subsequently selected for puromycin resistance. After drug selection, a pool of colonies was lysed, and UBR4 knockdown efficiency was determined using 5% SDS-PAGE and immunoblotting. A nonspecific band is indicated by the asterisk as a loading control. Independent immunoblot analysis with isolated colonies gave largely similar results (data not shown). (B) Pulse-chase analysis of X-nsP4, where X is Met (stabilizing), Arg (type 1 N-degron), or Tyr (type 2 N-degron), in cells deficient in UBR proteins, singly or in combination. See Fig. 1F for details. (C) Quantitation of the patterns shown in B using PhosphorImager.
FIG. 6.
Degradation kinetics of X-nsP4 bearing N-terminal His, Phe, Gly, Leu, and Thr in various UBR mutant cells. (A) Pulse-chase analysis of X-nsp4, where X is Met or Gly (stabilizing), His (type 1 N-degron), Leu or Phe (type 2 N-degron), or Thr (type3 N-degron), in cells deficient in UBR proteins, singly or in combination. (B) Quantitation of the patterns shown in A using PhosphorImager. S, stabilizing; 1/2, type 1/2 N-degron; 3, type 3 N-degron. (C) Ubiquitylation assay of X-nsP4 in rabbit reticulocyte lysate. Upper panel: X-nsP4 proteins (where X is Met, Arg, or Tyr) were expressed and translationally labeled with biotin using transcription-translation-coupled reticulocyte lysates with or without 2 mM dipeptide as indicated. The proteasome inhibitor MG132 (5 μM) was also present, to accumulate ubiquitylated proteins. After 30 min of incubation, ubiquitylated proteins were immunoprecipitated with anti-Ub antibody, followed by SDS-PAGE of precipitated proteins and detection of biotinylated proteins (see Materials and Methods). Lower panel: X-nsP4 proteins (where X is Met, Arg, or Tyr) were in vitro translated as above without MG132 and a portion of the reticulocyte lysates was loaded, followed by Western blotting with horseradish peroxidase-conjugated streptavidin.
FIG. 7.
HIV-1 integrase is an in vivo substrate of UBR box proteins. (A) Schematic diagram for a bicistronic mRNA producing EGFP and X-integrase (where X is Met, Gly, Arg, His, Phe, or Leu) used in B. (B) The steady-state level of X-integrase and EGFP, expressed in wild-type, _UBR1_−/− _UBR2_−/− _Luc_RNAi, and _UBR1_−/− _UBR2_−/− _UBR4_RNAi cells, was determined using immunoprecipitation and Western analysis with anti-integrase antibody. Cells were treated with MG132 (10 μM) or solvent (dimethyl sulfoxide [DMSO]). N-terminal amino acids of integrase are indicated on the top. Ig-H and Ig-L, mouse immunoglobulin G heavy and light chains, respectively. The expression of the reference protein EGFP was determined using immunoblotting with anti-GFP antibody for an input control. (C) A pcDNA3-based bicistronic construct expressing Ub-X-integrase and hrGFP used in D to F. Since hrGFP was poorly expressed in EFs, LacZ-V5 was coexpressed as a reference protein. (D) Pulse-chase analysis of Met- and Phe-integrases in wild-type and _UBR1_−/− _UBR2_−/− _UBR4_RNAi cells. Cells were labeled for 30 min with [35S]methionine/cysteine, followed by a chase for 0, 20, and 40 min in the presence of cycloheximide, preparation of extracts, immunoprecipitation with a combination of anti-integrase and anti-V5 antibodies, followed by SDS-PAGE and autoradiography. (E) Quantitation of the patterns shown in D using a PhosphorImager. (F) Degradation of Met- and Phe-integrases was analyzed using in vitro transcription/translation-coupled rabbit reticulocyte lysate in the presence of MG132 (5 μM) or dipeptide (2 mM) as indicated, and [35S]methionine-labeled proteins were analyzed by SDS-PAGE, followed by autoradiography. Met-integrase is indeed slightly stabilized by MG132 or the type 1 dipeptide Arg-Ala (but not by the type 1 dipeptide Phe-Ala), suggesting that a minor population of Met-integrase may be modified (e.g., oxidation) into an unknown structure that mimics a weak type 1 N-degron.
FIG. 8.
Lentiviral integrases and alphaviral RNA polymerases commonly bear type 2 N-degron. (A, B) Alignment of inferred N-terminal sequences of mature lentiviral integrases and nonlentiviral integrases (A) and alphaviral RNA polymerases (B). Conserved type 2 N-degrons are indicated by blue highlighting and an asterisk. Other conserved residues are indicated by orange highlighting. The protein sequences (accession number, organism) listed are: BIV_106 (P19560, bovine immunodeficiency virus isolate 106), EIAV_CL22 (P32542, equine infectious anemia virus clone CL22), FIV_TM2 (P31822, feline immunodeficiency virus isolate TM2), CAEV_CORK (P33459, caprine arthritis encephalitis virus strain CORK), OLV_SA_OMV (P16901, ovine lentivirus strain SA-OMVV), SRLV, (AAS18421, Small ruminant lentivirus), visna_KV17 (P35956, Visna lentivirus strain KV1772), HIV-1_HXB2 (P04585, HIV-1 HXB2 isolate), HIV-1_YU-2 (P35963, HIV-1 YU-2 isolate), HIV-2_GHAN (P18042, HIV-2 isolate GHANA-1), HIV-2_KR (Q74120, HIV-2 isolate KR), SIV_AGM155 (P27973, simian immunodeficiency virus AGM155 isolate), SIV_GRI-1 (Q02836, simian immunodeficiency virus isolate AGM), HumERK (CAA76879, human endogenous retrovirus K), MMTV (GNMVMM, mouse mammary tumor virus), SRV2 (AAD43256, simian retrovirus 2), WMSV (NP_041261, woolly monkey sarcoma virus), MPMV (GNLJMP, Mason-Pfizer monkey virus), STLV2 (CAA74901, simian T-lymphotropic virus 2), WEEV (P_818936, western equine encephalomyelitis virus), EEEV (S72349, eastern equine encephalitis virus), VEEV (AAD14556, Venezuelan equine encephalitis virus), SFV (NP_463457, Semliki forest virus), Mayaro (NP_579968, Mayaro virus), Sindbis (P03317, Sindbis virus), Sleeping_d (NP_740656, sleeping disease virus), Igbo_Ora (NP_740715, Igbo Ora virus), SPD (NP_740638, salmon pancreas disease virus), Aura (NP_819013, aura virus), Middelburg (P03318, Middleburg virus), and Ross_river (P13887, Ross River virus strain NB5092).