Synergistic recognition of an epigenetic DNA element by Pleiohomeotic and a Polycomb core complex - PubMed (original) (raw)
Synergistic recognition of an epigenetic DNA element by Pleiohomeotic and a Polycomb core complex
Adone Mohd-Sarip et al. Genes Dev. 2005.
Abstract
Polycomb response elements (PREs) are cis-acting DNA elements that mediate epigenetic gene silencing by Polycomb group (PcG) proteins. Here, we report that Pleiohomeotic (PHO) and a multiprotein Polycomb core complex (PCC) bind highly cooperatively to PREs. We identified a conserved sequence motif, named PCC-binding element (PBE), which is required for PcG silencing in vivo. PHO sites and PBEs function as an integrated DNA platform for the synergistic assembly of a repressive PHO/PCC complex. We termed this nucleoprotein complex silenceosome to reflect that the molecular principles underpinning its assemblage are surprisingly similar to those that make an enhanceosome.
Figures
Figure 1.
DNA templates and purified proteins. (A) Schematic representation of the Ubx locus. The BXD/PBX regulatory regions and bxd PRE and Ubx promoter are indicated. Map positions are according to Bender et al. (1983). The eight PHO sites within the core of the bxd PRE are indicated. Numbering is according to Fritsch et al. (1999) and Mahmoudi et al. (2003). For our binding studies, we used a short 50-bp DNA fragment harboring the PHO4 and PHO5 sites, referred to as PHO4/5-PRE. These strong PHO-binding elements are required for bxd PRE-directed silencing in vivo (Fritsch et al. 1999). (B) Schematic representation of the regulatory region of Abd-B. The parasegment-specific regulatory domains iab-5, iab-6, and iab-7, part of the iab-8, and the iab-7 PRE are indicated (Karch et al. 1985). A minimal iab-7 PRE harboring three PHO sites is used in both reconstituted DNA-binding and in vivo silencing studies (Mihaly et al. 1998). (C) Recombinant PHO, PHO lacking the PBD (ΔPBD: amino acids 1-148/169-520), PC, and PCC were immunopurified from extracts of baculovirus-infected Sf9 cells. The eluted proteins were resolved by SDS-PAGE and visualized by silver staining.
Figure 2.
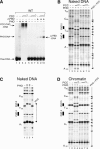
PHO and PCC bind synergistically to the bxd PRE. (A) PRE binding of PHO, ΔPBD and PCC was studied by bandshift assays. Binding reactions contained ∼5 nM PHO and/or ∼5-15 nM PCC, and radiolabeled PHO4/5-PRE. PRE binding of PHO and PCC was analyzed by primer extension DNaseI footprinting using either naked DNA (B,C) or chromatinized PHO4/5-PRE (D) templates. Binding reactions on naked DNA contained ∼20 nM (+) or 100 nM (‡) PHO and ∼20-60 nM PCC. Chromatin binding reactions contained ∼50 nM PHO and ∼20-60 nM PCC. DNaseI digestion ladders were analyzed by primer extension, resolved on a 6% polyacrylamide gel, and visualized by autoradiography. The positions of the PHO sites and footprinted areas are indicated. Closed arrowheads indicate DNaseI hypersensitive sites induced by protein binding.
Figure 3.
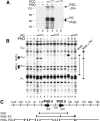
PCC subunits contact DNA. (A) UV cross-linking of PHO and PCC subunits to the PRE. Binding reactions contained a 32P-labeled, BrdU-substituted PHO4/5-PRE fragment in the absence or presence of the indicated proteins. After UV cross-linking, proteins were analyzed by SDS-PAGE and visualized by autoradiography. The relative positions of PSC, PH, PC, and PHO are indicated. (B) PRE binding of PHO and PC was analyzed by primer extension DNaseI footprinting as described in Figure 2. Reactions contained ∼20 or 80 nM PC. (C) Summary of the DNaseI footprinting patterns on PHO4/5-PRE.
Figure 4.
The PBE is required for PCC recruitment to the PRE. (A) Alignment of a selection of PHO sites and their flanking motifs from Mihaly et al. (1998), including the bxd PRE PHO sites 4 and 5 and iab-7 PHO sites. The PHO/YY1 consensus core (C) sequence GCCAT and the upstream (U) and downstream (D or PBE) flanking motifs are indicated. The U.mt, C.mt, and D.mt are indicated for PHO4 and PHO5. Corresponding D.mts were induced in the iab-7 PBEs (Fig. 5). The effects of mutations on PHO binding were tested by DNaseI footprinting on the PHO4/5 PRE (B) and by bandshift analysis (C). (D) Synergistic binding of PHO and PCC requires both PHO4 and PHO5 sites and both accompanying PBEs. Binding was assayed as described above using probes harboring the indicated mutations. (E) Model illustrating that cooperative DNA binding by PHO and PCC requires (1) at least two PHO sites, (2) their juxtaposed PBEs, and (3) direct protein-protein interactions between PHO and PCC. PHO sites and PBEs form an integrated platform for the synergistic assembly of a repressive PHO/PCC/PRE nucleoprotein complex. Details are discussed in the text.
Figure 5.
The PBE is required for PRE silencing in vivo. (A) Binding of PHO/PCC to the iab-7 PRE is abrogated when the PBEs are mutated (for sequences, see Fig. 4A), as revealed by DNaseI footprinting. (_B_-D) Mutations in the PBE impair PRE-directed PSS. PSS of mini-white by the minimal 260-bp wild-type iab-7 PRE or a mutant, PBE-less iab-7 PRE (PBEmt iab-7 PRE) was compared. Multiple independent lines were established harboring the mini-white transgene under control of either the wild-type or PBEmt iab-7 PRE. For each line, the eye color of homozygotes for the insertion was compared with that of heterozygotes. Representative examples are shown: (B) Homozygotes have a much darker eye color than their heterozygous siblings (P[w+]/P[w+]P[w+]/+), revealing the absence of PSS. (C) Homozygotes have much lighter eyes than heterozygotes ((P[w+]/P[w+][w+]/+), showing strong PSS. (D) The eye color of homozygotes is equal (P[w+]/P[w+] = P[w+]/+) to that of heterozygotes, revealing impaired PSS. (E) The number of lines exhibiting no PSS, impaired PSS, or strong PSS were scored and depicted in a graphical representation with the number of lines displaying a PSS phenotype expressed as percentage of the total number of lines analyzed on the _Y_-axis and the phenotype on the _X_-axis. Because the percentages refer to the numbers of independent lines, error bars reflecting the SEM are not applicable. χ2 test of statistical significance for bivariate tabular analysis confirmed the high significance of the difference in PSS frequency between lines harboring either the wild-type or PBEmt iab-7 PREs (χ2 = 12.3, p < 0.01).
Similar articles
- Architecture of a polycomb nucleoprotein complex.
Mohd-Sarip A, van der Knaap JA, Wyman C, Kanaar R, Schedl P, Verrijzer CP. Mohd-Sarip A, et al. Mol Cell. 2006 Oct 6;24(1):91-100. doi: 10.1016/j.molcel.2006.08.007. Mol Cell. 2006. PMID: 17018295 - Combgap contributes to recruitment of Polycomb group proteins in Drosophila.
Ray P, De S, Mitra A, Bezstarosti K, Demmers JA, Pfeifer K, Kassis JA. Ray P, et al. Proc Natl Acad Sci U S A. 2016 Apr 5;113(14):3826-31. doi: 10.1073/pnas.1520926113. Epub 2016 Mar 21. Proc Natl Acad Sci U S A. 2016. PMID: 27001825 Free PMC article. - The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing.
Brown JL, Fritsch C, Mueller J, Kassis JA. Brown JL, et al. Development. 2003 Jan;130(2):285-94. doi: 10.1242/dev.00204. Development. 2003. PMID: 12466196 - Polycomb silencing mechanisms and the management of genomic programmes.
Schwartz YB, Pirrotta V. Schwartz YB, et al. Nat Rev Genet. 2007 Jan;8(1):9-22. doi: 10.1038/nrg1981. Nat Rev Genet. 2007. PMID: 17173055 Review. - Polycomb response elements and targeting of Polycomb group proteins in Drosophila.
Müller J, Kassis JA. Müller J, et al. Curr Opin Genet Dev. 2006 Oct;16(5):476-84. doi: 10.1016/j.gde.2006.08.005. Epub 2006 Aug 17. Curr Opin Genet Dev. 2006. PMID: 16914306 Review.
Cited by
- Context-dependent role of Pho binding sites in Polycomb complex recruitment in Drosophila.
Brown JL, Price JD, Erokhin M, Kassis JA. Brown JL, et al. Genetics. 2023 Aug 9;224(4):iyad096. doi: 10.1093/genetics/iyad096. Genetics. 2023. PMID: 37216193 Free PMC article. - DNA binding by polycomb-group proteins: searching for the link to CpG islands.
Owen BM, Davidovich C. Owen BM, et al. Nucleic Acids Res. 2022 May 20;50(9):4813-4839. doi: 10.1093/nar/gkac290. Nucleic Acids Res. 2022. PMID: 35489059 Free PMC article. Review. - Distinct requirements for Pho, Sfmbt, and Ino80 for cell survival in Drosophila.
Elizarev P, Finkl K, Müller J. Elizarev P, et al. Genetics. 2021 Aug 26;219(1):iyab096. doi: 10.1093/genetics/iyab096. Genetics. 2021. PMID: 34849913 Free PMC article. - Polycomb group-mediated histone H2A monoubiquitination in epigenome regulation and nuclear processes.
Barbour H, Daou S, Hendzel M, Affar EB. Barbour H, et al. Nat Commun. 2020 Nov 23;11(1):5947. doi: 10.1038/s41467-020-19722-9. Nat Commun. 2020. PMID: 33230107 Free PMC article. Review. - Evolving Role of RING1 and YY1 Binding Protein in the Regulation of Germ-Cell-Specific Transcription.
Bajusz I, Henry S, Sutus E, Kovács G, Pirity MK. Bajusz I, et al. Genes (Basel). 2019 Nov 19;10(11):941. doi: 10.3390/genes10110941. Genes (Basel). 2019. PMID: 31752312 Free PMC article. Review.
References
- Bender W., Akam, M., Karch, F., Beachy, P., Peifer, M., Spierer, P., Lewis, E., and Hogness, D. 1983. Molecular genetics of the bithorax complex in Drosophila melanogaster. Science 221: 23-29. - PubMed
- Brown J.L., Mucci, D., Whiteley, M., Dirksen, M.L., and Kassis, J.A. 1998. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA-binding protein with homology to the transcription factor YY1. Mol. Cell 1: 1057-1064. - PubMed
- Brown J.L., Fritsch, C., Mueller, J., and Kassis, J.A. 2003. The Drosophila pho-like gene encodes a YY1-related DNA-binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development 130: 285-294. - PubMed
- Carey M. 1998. The enhanceosome and transcriptional synergy. Cell 92: 5-8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases