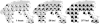Recovering the geographic origin of early modern humans by realistic and spatially explicit simulations - PubMed (original) (raw)
Recovering the geographic origin of early modern humans by realistic and spatially explicit simulations
Nicolas Ray et al. Genome Res. 2005 Aug.
Abstract
Most genetic and archeological evidence argue in favor of a recent and unique origin of modern humans in sub-Saharan Africa, but no attempt has ever been made at quantifying the likelihood of this model, relative to alternative hypotheses of human evolution. In this paper, we investigate the possibility of using multilocus genetic data to correctly infer the geographic origin of humans, and to distinguish between a unique origin (UO) and a multiregional evolution (ME) model. We introduce here an approach based on realistic simulations of the genetic diversity expected after an expansion process of modern humans into the Old World from different possible areas and their comparison to observed data. We find that the geographic origin of the expansion can be correctly recovered provided that a large number of independent markers are used, and that precise information on past demography and potential places of origins is available. In that case, it is also possible to unambiguously distinguish between a unique origin and a multiregional model of human evolution. Application to a real human data set of 377 STR markers tested in 22 populations points toward a unique but surprising North African origin of modern humans. We show that this result could be due to ascertainment bias in favor of markers selected to be polymorphic in Europeans. A new estimation modeling this bias explicitly reveals that East Africa is the most likely place of origin for modern humans.
Figures
Figure 1.
(A) Location, name, and size (within parentheses) of the 22 populations drawn from Rosenberg et al. (2002) for which genetic diversity was simulated. Arrows indicate the location of the four land bridges introduced to allow the complete colonization of the Old World and Oceania, since four samples (Orcadian, Sardinian, Japanese, and Papuan) are localized in islands. We imposed a low carrying capacity (K = 10) for demes located on these bridges to simulate a likely Founder Effect associated with the settlement of these islands. (B) Location of the 25 simulated origins (black dots). Open squares indicate the location of the 14 pseudo-observed origins. The four genetic regions defined in Rosenberg et al. (2002) are defined by the straight black lines. (C) Representation of relative carrying capacity values. (D) Representation of relative friction values. In C and D, darker colors indicate relatively higher carrying capacity or friction values.
Figure 2.
Demography and timeline of the different simulated evolutionary models. (A) Unique origin (UO) model. In this model, 30,000 generations ago, a small population (N = 100 genes) went through a demographic expansion after a first speciation event. Then, 4000 generations ago, a range expansion followed a bottleneck of 10 generations to mimic a second speciation event. The large population preceding the speciation and range expansion can be considered to be a large subdivided population. (B) Multiregional evolution (ME) model. As in A, a small population went through a speciation event and instantaneously colonized the three continents 30,000 generations ago. For 26,000 generations the continents harbored relatively large populations and exchanged occasional migrants (see Table 1 for continent population sizes and migration rates under different scenarios). Then, 4000 generations ago, three range expansions were initiated from the three different origins shown in C.
Figure 3.
Relative frequencies of correct assignment for 25 simulated origins and for different numbers of available STR loci. The black area represents the proportion of simulations for which the origin of demographic expansion was correctly recovered from the R90 statistic. The frequencies of correct assignment for the complete set of simulations are given in Table 2.
Figure 4.
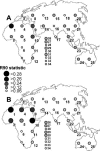
Values of the R90 statistic for 25 UO scenarios (nos. 1–25) and nine ME scenarios (nos. 26–34) computed from the Rosenberg22 data set under the unbiased (A) and biased (B) simulated data sets. Exact values of the R90 statistics are reported in Supplemental Table 2.
Similar articles
- Estimating relative population sizes from simulated data sets and the question of greater African effective size.
Eller E. Eller E. Am J Phys Anthropol. 2001 Sep;116(1):1-12. doi: 10.1002/ajpa.1096. Am J Phys Anthropol. 2001. PMID: 11536112 - Origins and affinities of modern humans: a comparison of mitochondrial and nuclear genetic data.
Jorde LB, Bamshad MJ, Watkins WS, Zenger R, Fraley AE, Krakowiak PA, Carpenter KD, Soodyall H, Jenkins T, Rogers AR. Jorde LB, et al. Am J Hum Genet. 1995 Sep;57(3):523-38. doi: 10.1002/ajmg.1320570340. Am J Hum Genet. 1995. PMID: 7668280 Free PMC article. - Craniometric variation, genetic theory, and modern human origins.
Relethford JH, Harpending HC. Relethford JH, et al. Am J Phys Anthropol. 1994 Nov;95(3):249-70. doi: 10.1002/ajpa.1330950302. Am J Phys Anthropol. 1994. PMID: 7856764 Review. - GM haplotype diversity of 82 populations over the world suggests a centrifugal model of human migrations.
Dugoujon JM, Hazout S, Loirat F, Mourrieras B, Crouau-Roy B, Sanchez-Mazas A. Dugoujon JM, et al. Am J Phys Anthropol. 2004 Oct;125(2):175-92. doi: 10.1002/ajpa.10405. Am J Phys Anthropol. 2004. PMID: 15365983 - Genetic perspectives on human origins and differentiation.
Harpending H, Rogers A. Harpending H, et al. Annu Rev Genomics Hum Genet. 2000;1:361-85. doi: 10.1146/annurev.genom.1.1.361. Annu Rev Genomics Hum Genet. 2000. PMID: 11701634 Review.
Cited by
- Process-explicit models reveal the structure and dynamics of biodiversity patterns.
Pilowsky JA, Colwell RK, Rahbek C, Fordham DA. Pilowsky JA, et al. Sci Adv. 2022 Aug 5;8(31):eabj2271. doi: 10.1126/sciadv.abj2271. Epub 2022 Aug 5. Sci Adv. 2022. PMID: 35930641 Free PMC article. Review. - Pan-Africanism vs. single-origin of Homo sapiens: Putting the debate in the light of evolutionary biology.
Meneganzin A, Pievani T, Manzi G. Meneganzin A, et al. Evol Anthropol. 2022 Jul;31(4):199-212. doi: 10.1002/evan.21955. Epub 2022 Jul 18. Evol Anthropol. 2022. PMID: 35848454 Free PMC article. Review. - Addressing alpine plant phylogeography using integrative distributional, demographic and coalescent modeling.
Larsson DJ, Pan D, Schneeweiss GM. Larsson DJ, et al. Alp Bot. 2022;132(1):5-19. doi: 10.1007/s00035-021-00263-w. Epub 2021 Jul 29. Alp Bot. 2022. PMID: 35368907 Free PMC article. Review. - Evaluating Causes of Current Genetic Gradients of Modern Humans of the Iberian Peninsula.
Ferreiro D, Núñez-Estévez B, Canedo M, Branco C, Arenas M. Ferreiro D, et al. Genome Biol Evol. 2021 Apr 5;13(4):evab071. doi: 10.1093/gbe/evab071. Genome Biol Evol. 2021. PMID: 33837782 Free PMC article. - SPLATCHE3: simulation of serial genetic data under spatially explicit evolutionary scenarios including long-distance dispersal.
Currat M, Arenas M, Quilodràn CS, Excoffier L, Ray N. Currat M, et al. Bioinformatics. 2019 Nov 1;35(21):4480-4483. doi: 10.1093/bioinformatics/btz311. Bioinformatics. 2019. PMID: 31077292 Free PMC article.
References
- Brauer, G., Collard, M., and Stringer, C. 2004. On the reliability of recent tests of the Out of Africa hypothesis for modern human origins. Anat. Rec. 279A: 701–707. - PubMed
- Cavalli-Sforza, L.L. and Feldman, M.W. 2003. The application of molecular genetic approaches to the study of human evolution. Nat. Genet. 33: 266–275. - PubMed
- Cavalli-Sforza, L.L., Menozzi, P., and Piazza, A. 1994. The history and geography of human genes. Princeton University Press, Princeton, NJ.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous