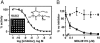Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry - PubMed (original) (raw)
Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry
Graham Simmons et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Severe acute respiratory syndrome (SARS) is caused by an emergent coronavirus (SARS-CoV), for which there is currently no effective treatment. SARS-CoV mediates receptor binding and entry by its spike (S) glycoprotein, and infection is sensitive to lysosomotropic agents that perturb endosomal pH. We demonstrate here that the lysosomotropic-agent-mediated block to SARS-CoV infection is overcome by protease treatment of target-cell-associated virus. In addition, SARS-CoV infection was blocked by specific inhibitors of the pH-sensitive endosomal protease cathepsin L. A cell-free membrane-fusion system demonstrates that engagement of receptor followed by proteolysis is required for SARS-CoV membrane fusion and indicates that cathepsin L is sufficient to activate membrane fusion by SARS-CoV S. These results suggest that SARS-CoV infection results from a unique, three-step process: receptor binding and induced conformational changes in S glycoprotein followed by cathepsin L proteolysis within endosomes. The requirement for cathepsin L proteolysis identifies a previously uncharacterized class of inhibitor for SARS-CoV infection.
Figures
Fig. 1.
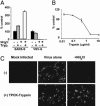
Effect of trypsin on SARS-CoV infection. (A) Trypsin treatment bypasses ammonium chloride inhibition. HIV-luc(SARS S) or HIV-luc(VSV-G) were bound to mock (black and gray bars) or ammonium chloride-treated (third set of bars and white bars) 293T/ACE2 cells. The cells were incubated with either PBS (black bars and third set of bars) or TPCK-trypsin (gray and white bars). The results are presented as a percentage of no-ammonium-chloride (NH4Cl), no-trypsin (Tryp.) controls (≈4,000 and 10,000 RLU for SARS S and VSV-G, respectively) and represent the means of samples run in triplicate (±SD). Similar results were seen in two subsequent assays. (B) Trypsin pretreatment of S protein inactivates infectivity. HIV-luc(SARS S) infection of 293T/ACE2 cells was assessed as luciferase activity, presented as a percentage of no-trypsin control (≈40,000 RLU). The results represent the means of samples run in triplicate (±SD). (C) Trypsin treatment bypasses ammonium chloride inhibition of SARS-CoV. Mock- (Center) or 25 mM ammonium chloride-pretreated (Right) Vero E6 cells were spin-infected with replication-competent SARS-CoV at a multiplicity of infection of 0.5 and incubated with either DMEM (Upper) or DMEM containing TPCK-trypsin (Lower). After 48 h, the cells were immunostained for S protein.
Fig. 2.
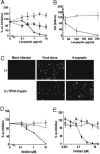
Protease-inhibitor sensitivity. (A) Leupeptin inhibits S protein-mediated infection. The 293T cells were preincubated with leupeptin and challenged with HIV-luc SARS S (solid line, ♦), VSV-G (dashed line, ▪), or MLV-Ampho (dotted line, ▴). The results are presented as a percentage of infection of untreated cells (≈3,000 RLU) for each envelope) and represent the means of samples run in triplicate (±SD). Similar results were seen in two subsequent assays. (B) Leupeptin inhibits replication-competent SARS-CoV infection. Cells were either preincubated with leupeptin for 1 h and then exposed to virus for 3 h in the continued presence of leupeptin (solid line) or exposed to virus for 3 h and incubated for an additional 4 h with leupeptin (dashed line). At 3 days postexposure, the supernatant was analyzed for nucleoprotein by ELISA. The results are expressed as OD and represent the means of samples run in triplicate (±SD). Similar results were seen in a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium cytoxicity assay. (C) Trypsin treatment bypasses leupeptin inhibition of live SARS-CoV. Mock- (Center) or 500 μg/ml leupeptin-pretreated (Right) Vero E6 cells were spin-infected with replication-competent SARS-CoV at a multiplicity of infection of 0.5 and incubated with either DMEM (Upper) or DMEM containing TPCK-trypsin (Lower). After 48 h, the cells were immunostained for S protein. (D) E64c blocks SARS-CoV S protein-mediated entry. The 293T cells were preincubated with E64c (solid lines) or aprotinin (dashed lines) and challenged with HIV-luc SARS S (black lines) or VSV-G (gray lines). The results are presented as a percentage of infection of untreated cells (≈1,500 RLU for VSV-G and 6,000 RLU for SARS S) and represent the means of samples run in triplicate (±SD). Similar results were seen in two additional experiments. (E) Z-lll-FMK inhibits S protein-mediated infection. Vero E6 cells were preincubated with Z-lll-FMK (solid lines) or CA-074 (dashed lines) and then challenged with HIV-luc SARS S (black lines) or VSV-G (gray lines). The results are presented as a percentage of infection of untreated cells (≈15,000 RLU for VSV-G and 20,000 RLU for SARS S) and represent the means of samples run in triplicate (±SD). Similar results were seen on 293T and 293T/ACE2 cells.
Fig. 3.
Cathepsin-L-specific inhibitor blocks infection. (A) MDL28170 inhibits CTSL activity with an IC50 of 2.5 nM. A 1,000-compound library was screened for inhibitors of CTSL activity (Inset, bottom left). MDL28170 (Inset, top right) was found to be a potent inhibitor. The compound library was screened against several other cathepsins, including CTSB, with no hits. The activity of MDL28170 was confirmed in an in vitro CTSL-cleavage assay (inhibition curve). (B) MDL28170 inhibits S protein-mediated infection. The 293T cells were preincubated with MDL28170 and challenged with HIV-luc SARS S (solid line) or VSV-G (dashed line). The results are presented as a percentage of infection of untreated cells (≈100,000 RLU for VSV-G and 20,000 RLU for SARS S) and represent the means of samples run in triplicate (±SD). Similar results were seen on Vero E6 and 293T/ACE2 cells.
Fig. 4.
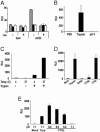
S protein-mediated intervirion fusion. (A) Intervirion fusion requires ACE2 and S protein. Bald or ACE2 particles encoding luciferase (x axis) were incubated with particles encoding GFP (SARS S and ASLV-A envelope, gray bars; SARS S alone, black bars; or ASLV-A envelope alone, white bars). Virions were mixed and used to infect HeLa/Tva cells that had been pretreated with medium in the presence and absence of leupeptin (Leu) (20 μg/ml). Intervirion fusion was measured as luciferase activity 48 h postinfection. Results represent the means of samples run in triplicate (±SD). (B) Trypsin cleavage promotes fusion mediated by S protein. Intervirion fusion between HIV-luc(ACE2) and HIV-gfp(SARS S/ASLV-A) treated with TPCK-trypsin (10 μg/ml) for 10 min at 25°C or pulsed at pH 5.0 was quantified by luciferase activity 48 h postinfection of HeLa/Tva cells pretreated with leupeptin. The results represent the means of samples run in triplicate (±SD). Mixtures of HIV-gfp(SARS S), HIV-gfp(ASLV-A), and HIV-luc(ACE2) could not be activated by trypsin cleavage, suggesting that S and ASLV-A envelope are required to be incorporated into the same particle in order for transduction of target cells by fused particles. (C) Receptor interactions at elevated temperature are required before trypsin cleavage. HIV-luc(ACE2) and HIV-GFP(SARS S/ASLV-A) particles were mixed and incubated at 4°C to allow binding. Samples were then incubated at the noted temperatures. TPCK-trypsin digestion was carried out at 4°C for 15 min. The results represent the means of samples run in quadruplicate (±SD). Similar results were observed in two additional experiments. Temp., temperature. (D) CTSL enhances intervirion fusion. HIV-luc(ACE2) and HIV-GFP(SARS S/ASLV-A) particles were mixed and incubated for 10 min at 25°C with preactivated CTSB (at pH 5.0), CTSL (at pH 6.0), CTSL buffer alone (at pH 6.0), or TPCK-trypsin (at pH 7.0). The mixed virus was used to infect HeLa/Tva cells pretreated with leupeptin. The results represent the means of samples run in quadruplicate (±SD). Similar results were observed in two subsequent experiments. (E) Acidic conditions are required for CTSL-mediated S protein activation. HIV-luc(ACE2) and HIV-GFP(SARS S/ASLV-A) particles were mixed and adjusted to various pHs and CTSL was added. After neutralization of acid conditions, the mixed virus was used to infect HeLa/Tva cells pretreated with leupeptin. The results represent the means of samples run in quadruplicate (±SD). Tryp, trypsin. Similar results were observed in an additional experiment.
Similar articles
- Amiodarone alters late endosomes and inhibits SARS coronavirus infection at a post-endosomal level.
Stadler K, Ha HR, Ciminale V, Spirli C, Saletti G, Schiavon M, Bruttomesso D, Bigler L, Follath F, Pettenazzo A, Baritussio A. Stadler K, et al. Am J Respir Cell Mol Biol. 2008 Aug;39(2):142-9. doi: 10.1165/rcmb.2007-0217OC. Epub 2008 Feb 28. Am J Respir Cell Mol Biol. 2008. PMID: 18314540 - SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells.
Huang IC, Bosch BJ, Li F, Li W, Lee KH, Ghiran S, Vasilieva N, Dermody TS, Harrison SC, Dormitzer PR, Farzan M, Rottier PJ, Choe H. Huang IC, et al. J Biol Chem. 2006 Feb 10;281(6):3198-203. doi: 10.1074/jbc.M508381200. Epub 2005 Dec 8. J Biol Chem. 2006. PMID: 16339146 Free PMC article. - Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry.
Qiu Z, Hingley ST, Simmons G, Yu C, Das Sarma J, Bates P, Weiss SR. Qiu Z, et al. J Virol. 2006 Jun;80(12):5768-76. doi: 10.1128/JVI.00442-06. J Virol. 2006. PMID: 16731916 Free PMC article. - Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research.
Simmons G, Zmora P, Gierer S, Heurich A, Pöhlmann S. Simmons G, et al. Antiviral Res. 2013 Dec;100(3):605-14. doi: 10.1016/j.antiviral.2013.09.028. Epub 2013 Oct 8. Antiviral Res. 2013. PMID: 24121034 Free PMC article. Review. - SARS coronavirus anti-infectives.
Tong TR. Tong TR. Recent Pat Antiinfect Drug Discov. 2006 Nov;1(3):297-308. doi: 10.2174/157489106778777637. Recent Pat Antiinfect Drug Discov. 2006. PMID: 18221155 Review.
Cited by
- Potential of Flavonoid-Inspired Phytomedicines against COVID-19.
Ngwa W, Kumar R, Thompson D, Lyerly W, Moore R, Reid TE, Lowe H, Toyang N. Ngwa W, et al. Molecules. 2020 Jun 11;25(11):2707. doi: 10.3390/molecules25112707. Molecules. 2020. PMID: 32545268 Free PMC article. - Cathepsins L and B target HIF1α for oxygen-independent proteolytic cleavage.
Stuart S, Tarade D, Ohh M. Stuart S, et al. Sci Rep. 2024 Jun 26;14(1):14799. doi: 10.1038/s41598-024-65537-9. Sci Rep. 2024. PMID: 38926538 Free PMC article. - Human ACE2 receptor polymorphisms and altered susceptibility to SARS-CoV-2.
Suryamohan K, Diwanji D, Stawiski EW, Gupta R, Miersch S, Liu J, Chen C, Jiang YP, Fellouse FA, Sathirapongsasuti JF, Albers PK, Deepak T, Saberianfar R, Ratan A, Washburn G, Mis M, Santhosh D, Somasekar S, Hiranjith GH, Vargas D, Mohan S, Phalke S, Kuriakose B, Antony A, Ustav M Jr, Schuster SC, Sidhu S, Junutula JR, Jura N, Seshagiri S. Suryamohan K, et al. Commun Biol. 2021 Apr 12;4(1):475. doi: 10.1038/s42003-021-02030-3. Commun Biol. 2021. PMID: 33846513 Free PMC article. - Alpha-1-antitrypsin: A possible host protective factor against Covid-19.
de Loyola MB, Dos Reis TTA, de Oliveira GXLM, da Fonseca Palmeira J, Argañaraz GA, Argañaraz ER. de Loyola MB, et al. Rev Med Virol. 2021 Mar;31(2):e2157. doi: 10.1002/rmv.2157. Epub 2020 Aug 26. Rev Med Virol. 2021. PMID: 32844538 Free PMC article. Review. - Chinese hamster ovary cell lines selected for resistance to ebolavirus glycoprotein mediated infection are defective for NPC1 expression.
Haines KM, Vande Burgt NH, Francica JR, Kaletsky RL, Bates P. Haines KM, et al. Virology. 2012 Oct 10;432(1):20-8. doi: 10.1016/j.virol.2012.05.018. Epub 2012 Jun 21. Virology. 2012. PMID: 22726751 Free PMC article.
References
- Rota, P. A., Oberste, M. S., Monroe, S. S., Nix, W. A., Campagnoli, R., Icenogle, J. P., Penaranda, S., Bankamp, B., Maher, K., Chen, M. H., et al. (2003) Science 300, 1394-1399. - PubMed
- Guan, Y., Zheng, B. J., He, Y. Q., Liu, X. L., Zhuang, Z. X., Cheung, C. L., Luo, S. W., Li, P. H., Zhang, L. J., Guan, Y. J., et al. (2003) Science 302, 276-278. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI43455/AI/NIAID NIH HHS/United States
- R21 AI058701/AI/NIAID NIH HHS/United States
- R01 AI043455/AI/NIAID NIH HHS/United States
- R21 AI 058701/AI/NIAID NIH HHS/United States
- R21 AI059172/AI/NIAID NIH HHS/United States
- U54 AI057168/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous