Analysis of the biochemical mechanisms for the endocrine actions of fibroblast growth factor-23 - PubMed (original) (raw)
Analysis of the biochemical mechanisms for the endocrine actions of fibroblast growth factor-23
Xijie Yu et al. Endocrinology. 2005 Nov.
Abstract
Fibroblast growth factor (FGF)-23 has emerged as an endocrine regulator of phosphate and of vitamin D metabolism. It is produced in bone and, unlike other FGFs, circulates in the bloodstream to ultimately regulate phosphate handling and vitamin D production in the kidney. Presently, it is unknown which of the seven principal FGF receptors (FGFRs) transmits FGF23 biological activity. Furthermore, the molecular basis for the endocrine mode of FGF23 action is unclear. Herein, we performed surface plasmon resonance and mitogenesis experiments to comprehensively characterize receptor binding specificity. Our data demonstrate that FGF23 binds and activates the c splice isoforms of FGFR1-3, as well as FGFR4, but not the b splice isoforms of FGFR1-3. Interestingly, highly sulfated and longer glycosaminoglycan (GAG) species were capable of promoting FGF23 mitogenic activity. We also show that FGF23 induces tyrosine phosphorylation and inhibits sodium-phosphate cotransporter Npt2a mRNA expression using opossum kidney cells, a model kidney proximal tubule cell line. Removal of cell surface GAGs abolishes the effects of FGF23, and exogenous highly sulfated GAG is capable of restoring FGF23 activity, suggesting that proximal tubule cells naturally express GAGs that are permissive for FGF23 action. We propose that FGF23 signals through multiple FGFRs and that the unique endocrine actions of FGF23 involve escape from FGF23-producing cells and circulation to the kidney, where highly sulfated GAGs most likely act as cofactors for FGF23 activity. Our biochemical findings provide important insights into the molecular mechanisms by which dysregulated FGF23 signaling leads to disorders of hyper- and hypophosphatemia.
Figures
Fig. 1
Analysis of FGF23-FGFR binding by SPR. SPR sensorgrams illustrating FGF23 binding to FGFR1c (A), FGFR2c (B), FGFR3c (C), and FGFR4 (D). Analyte concentrations are colored as follows: 25 nM in gray, 50 nM in blue, 100 nM in red, 200 nM in green, 400 nM in violet, 800 nM in brown, and 1000 nM in gold. The biosensor chip response is indicated on the y-axis (RU) as a function of time (x-axis). Binding experiments were performed at 25 C.
Fig. 2
Activation of FGFRs by FGF23 assessed by cell proliferation. A, BaF3 cell lines each expressing one of the FGFR subtypes (FGFR1b, 1c, 2b, 2c, 3b, 3c, and 4) were treated with FGF23 (100 nM) and a preparation of unbleached heparin (2 µg/ml). FGF23 activated the c isoforms of FGFR1-3 and FGFR4. It showed negligible activity toward the b isoforms of FGFR1-3. B, FGF1 (5 nM) activated all FGFR isoforms. The results are plotted as percentage of control (no treatment) for each cell line. a, P < 0.05 vs. control (CTL).
Fig. 3
Activation of FGFR1c by FGF23 in the presence of sulfated GAGs. A, BaF3 cells expressing FGFR1c were treated with FGF23 (100 nM) with or without addition of heparin (2 µg/ml). The mitogenic activity of FGF23 was significantly increased by fully sulfated N, _O_-SH, _O_-DS, _O_-CS, and _O_-HA. In contrast, highly sulfated polysaccharides, as well as HS-M, did not synergize with FGF23 [a, P < 0.001 vs. CTL; b, P < 0.001 vs. FGF23 alone; c, P < 0.001 vs. respective heparin derivative alone]. B, FGF1 (5 nM) activated FGFR1c regardless of the sugar backbone of the GAG (P < 0.0001, vs. CTL, FGF1 alone, and respective heparin derivative alone).
Fig. 4
Dose dependence of FGFR1c activation by FGF23 and N, _O_-SH. A, BaF3 cells expressing FGFR1c were treated with increasing doses of FGF23 in the presence of 2 µg/ml N, _O_-SH. Mitogenic activity of FGF23 showed dose dependence, with a threshold dose of approximately 25 nM and an ED50 of approximately 50 nM (c, P < 0.0001 vs. N, _O_-SH alone). B, Increasing doses of N, _O_-SH resulted in increasing activity of FGF23 [a, P < 0.001 vs. control (CTL); b, P < 0.001 vs. FGF23 alone; c, P < 0.001 vs. heparin alone].
Fig. 5
FGF23 activity correlates with dp of heparin. BaF3 cells expressing FGFR1c were treated with FGF23 (100 nM) and heparins with increasing dp. Synergism of heparins with FGF23 became evidence at dp10 [P < 0.001 vs. control (CTL), treatment with dp20 alone]. A doubling of FGF23 activity was observed with heparins of dp in the range of 14–16 (P < 0.0001 vs. control).
Fig. 6
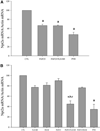
Effect of FGF23 on cellular tyrosine kinase phosphorylation. OK cells were treated for 15 min with either vehicle (CTL), PTH (100 nM), or FGF23 (100 nM) with or without 2 µg/ml N,O-SH, and the effects on cellular tyrosine kinase phosphorylation were examined by ELISA. FGF23 significantly increased cellular tyrosine kinase independently of exogenous fully sulfonated N, _O_-SH [P < 0.0001 vs. control (CTL)]. As expected, PTH had no effect on tyrosine kinase phosphorylation in OK cells. Data shown compared with vehicle control (a, P < 0.0001 vs. control). NS, not significant.
Fig. 7
Effect of FGF23 on NPT2a mRNA expression. A, OK cells were treated over 24 h with either vehicle (CTL), PTH (100 nM), or FGF23 (100 nM) with or without 2 µg/ml N,O-SH, and the effects on Npt2a mRNA were examined. FGF23 robustly decreased Npt2a mRNA expression, and its effect did not require N, _O_-SH [P < 0.001 vs. control (CTL)]. PTH, included as a positive control, had a similar effect. B, OK cells were treated with sodium chlorate (50 mM) to block sulfation of GAGs. PTH or FGF23 was added with or without 2 µg/ml N, O_-SH or HS-M. In this condition, the effect of FGF23 on Npt2a mRNA became sensitive to exogenous_N, _O_-SH. a, P < 0.05 vs. CTL; b, P < 0.05 vs. FGF23 alone; c,P < 0.05 vs. heparin alone.
Similar articles
- Convergent Signaling Pathways Regulate Parathyroid Hormone and Fibroblast Growth Factor-23 Action on NPT2A-mediated Phosphate Transport.
Sneddon WB, Ruiz GW, Gallo LI, Xiao K, Zhang Q, Rbaibi Y, Weisz OA, Apodaca GL, Friedman PA. Sneddon WB, et al. J Biol Chem. 2016 Sep 2;291(36):18632-42. doi: 10.1074/jbc.M116.744052. Epub 2016 Jul 18. J Biol Chem. 2016. PMID: 27432882 Free PMC article. - Regulation of renal phosphate transport by FGF23 is mediated by FGFR1 and FGFR4.
Gattineni J, Alphonse P, Zhang Q, Mathews N, Bates CM, Baum M. Gattineni J, et al. Am J Physiol Renal Physiol. 2014 Feb 1;306(3):F351-8. doi: 10.1152/ajprenal.00232.2013. Epub 2013 Nov 20. Am J Physiol Renal Physiol. 2014. PMID: 24259513 Free PMC article. - FGF23 decreases renal NaPi-2a and NaPi-2c expression and induces hypophosphatemia in vivo predominantly via FGF receptor 1.
Gattineni J, Bates C, Twombley K, Dwarakanath V, Robinson ML, Goetz R, Mohammadi M, Baum M. Gattineni J, et al. Am J Physiol Renal Physiol. 2009 Aug;297(2):F282-91. doi: 10.1152/ajprenal.90742.2008. Epub 2009 Jun 10. Am J Physiol Renal Physiol. 2009. PMID: 19515808 Free PMC article. - Pleiotropic Actions of FGF23.
Erben RG. Erben RG. Toxicol Pathol. 2017 Oct;45(7):904-910. doi: 10.1177/0192623317737469. Epub 2017 Nov 2. Toxicol Pathol. 2017. PMID: 29096595 Free PMC article. Review. - Bone-kidney axis in systemic phosphate turnover.
Razzaque MS. Razzaque MS. Arch Biochem Biophys. 2014 Nov 1;561:154-8. doi: 10.1016/j.abb.2014.06.031. Epub 2014 Jul 2. Arch Biochem Biophys. 2014. PMID: 24997362 Review.
Cited by
- Molecular mechanisms of fibroblast growth factor signaling in physiology and pathology.
Belov AA, Mohammadi M. Belov AA, et al. Cold Spring Harb Perspect Biol. 2013 Jun 1;5(6):a015958. doi: 10.1101/cshperspect.a015958. Cold Spring Harb Perspect Biol. 2013. PMID: 23732477 Free PMC article. Review. - Convergent Signaling Pathways Regulate Parathyroid Hormone and Fibroblast Growth Factor-23 Action on NPT2A-mediated Phosphate Transport.
Sneddon WB, Ruiz GW, Gallo LI, Xiao K, Zhang Q, Rbaibi Y, Weisz OA, Apodaca GL, Friedman PA. Sneddon WB, et al. J Biol Chem. 2016 Sep 2;291(36):18632-42. doi: 10.1074/jbc.M116.744052. Epub 2016 Jul 18. J Biol Chem. 2016. PMID: 27432882 Free PMC article. - The role of fibroblast growth factor 23 in regulation of phosphate balance.
Wilson R, Mukherjee-Roy N, Gattineni J. Wilson R, et al. Pediatr Nephrol. 2024 Dec;39(12):3439-3451. doi: 10.1007/s00467-024-06395-5. Epub 2024 Jun 14. Pediatr Nephrol. 2024. PMID: 38874635 Review. - Klotho in cardiovascular disease: Current and future perspectives.
Donate-Correa J, Martín-Núñez E, Mora-Fernández C, Muros-de-Fuentes M, Pérez-Delgado N, Navarro-González JF. Donate-Correa J, et al. World J Biol Chem. 2015 Nov 26;6(4):351-7. doi: 10.4331/wjbc.v6.i4.351. World J Biol Chem. 2015. PMID: 26629318 Free PMC article. Review. - FGF23-mediated regulation of systemic phosphate homeostasis: is Klotho an essential player?
Razzaque MS. Razzaque MS. Am J Physiol Renal Physiol. 2009 Mar;296(3):F470-6. doi: 10.1152/ajprenal.90538.2008. Epub 2008 Nov 19. Am J Physiol Renal Physiol. 2009. PMID: 19019915 Free PMC article. Review.
References
- Johnson DE, Williams LT. Structural and functional diversity in the FGF receptor multigene family. Adv Cancer Res. 1993;60:1–41. - PubMed
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–15297. - PubMed
- The ADHR Consortium. Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat Genet. 2000;26:345–348. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL052622/HL/NHLBI NIH HHS/United States
- DK063934/DK/NIDDK NIH HHS/United States
- R01 HL052622/HL/NHLBI NIH HHS/United States
- R01 HL062244/HL/NHLBI NIH HHS/United States
- R01 GM038060/GM/NIGMS NIH HHS/United States
- R01 DE013686/DE/NIDCR NIH HHS/United States
- R01 DK063934/DK/NIDDK NIH HHS/United States
- P01 HD039952/HD/NICHD NIH HHS/United States
- HL062244/HL/NHLBI NIH HHS/United States
- HD39952/HD/NICHD NIH HHS/United States
- DE13686/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous